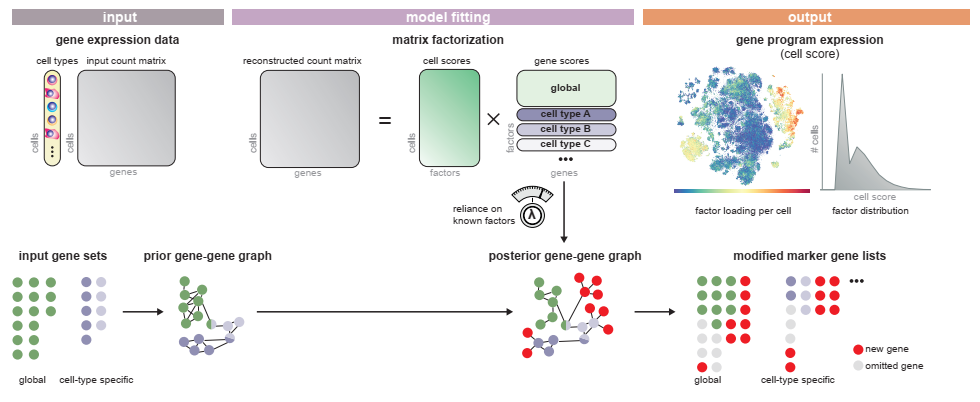
Overview
SPECTRA takes in a single cell gene expression matrix, cell type annotations, and a set of pathway annotations to fit the data to.
If you use Spectra please cite our preprint on bioRxiv.
gene_set_annotations = {
"global": {"global_ifn_II_response" : ["CIITA", "CXCL10", ...] , "global_MHCI": ["HLA-A". "HLA-B", "HLA-C"] },
"CD8_T": {"CD8_exhaustion" : ["CXCL13", "LAG3", "CD39", ...]},
...
}
Alternatively, SPECTRA can take in a set of adjacency matrices representing networks for each cell type:
gene_set_annotations = {
"global": adjacency_matrix1,
"CD8_T": adjacency_matrix2,
...
}
Installation
A pypi package will be available soon. For installation, you can add spectra to pip:
pip install git+https://github.com/dpeerlab/spectra
Tutorial
We provide a full tutorial how to run the basic Spectra model here:
You can use log1p transformed median library size normalized data. For leukocyte data, we recommend using scran normalization. We provide a tutorial.
scRNAseq Knowledge Base
Check out our scRNAseq knowledge base Cytopus 🐙 to retrieve Spectra input gene sets adapted to the cell type composition in your data.
Short Tutorial
We start by importing spectra. The easiest way to run spectra is to use the est_spectra function in the spectra module, as shown below. The default behavior is to set the number of factors equal to the number of gene sets plus one. However, this can be modified by passing an integer e.g. L = 20 as an argument to the function or a dictionary that maps cell type to an integer per cell type. We provide a method for estimating the number of factors directly from the data by bulk eigenvalue matching analysis, which is detailed further below.
from spectra import spectra as spc
model = spc.est_spectra(adata = adata, gene_set_dictionary = gene_set_dictionary, cell_type_key = "cell_type", use_highly_variable = True, lam = 0.01)
This function stores four important quantities in the AnnData, in addition to returning a fitted model object. Factors are the scores that tell you how much each gene contributes to each factor, while markers is an array of genes with top scores for every factor. Cell scores are similarly the score of each factor for every cell. Finally, vocab is a boolean array that is True for genes that were used while fitting the model - note that this quantity is only added to the AnnData when highly_variable is set to True.
factors = adata.uns['SPECTRA_factors'] # factors x genes matrix that tells you how important each gene is to the resulting factors
markers = adata.uns['SPECTRA_markers'] # factors x K [where K is a user specified number] list of K top markers per factor
cell_scores = adata.obsm['SPECTRA_cell_scores'] # cells x factors matrix of cell scores
vocab = adata.var['spectra_vocab'] # boolean matrix of size # of genes that indicates the set of genes used to fit spectra
We also provide an approach to label the factors by their Szymkiewicz–Simpson overlap coefficient with the input gene sets. Each factors receives the label of the input gene set with the highest overlap coefficient, given that it the overlap coefficient is greater than the threshold defined in 'overlap_threshold'. Ties in the overlap coefficient by gene set size, selecting the label of the bigger gene set (because smaller gene sets might get bigger overlap coefficients by chance).
We provide a pandas.DataFrame indicating the overlap coefficients for each input gene set with each factor's marker genes. The index of this dataframe contains the index of each factor, assigned label as well as the cell type specificity for each factor in the format:
['index' + '-X-' + 'cell type specificity' + '-X-' + 'assigned label', ...]
We use '-X-' as a unique seperator to make string splitting and retrieval of the different components of the index easier.
adata.uns['SPECTRA_overlap']
Accessing model parameters
To access finer grained information about the model fit, we can look at the attributes of the model object directly. Model parameters can be accessed with functions associated with the model object
model.return_eta_diag()
model.return_cell_scores()
model.return_factors()
model.return_eta()
model.return_rho()
model.return_kappa()
model.return_gene_scalings()
Apart from cell scores and factors, we can also retrive a number of other parameters this way that are not by default added to the AnnData. Eta diag is the diagonal of the fitted factor-factor interaction matrix; however, its interpretation is that it measures the extent to which each factor is influenced by the prior information. In practice many of these values are zero, indicating that they are estimated without bias introduced by the annotation set. Eta is the full set of factor-factor interaction matrices, whose off diagonals measure the extent to which factors share the same genes. Rho and kappa are parameters that control the background rate of non-edges and edges respectively. These can be fixed throughout training (default) or estimated from the data by providing rho = None or kappa = None to the est_spectra() function or to model.train(). Finally gene scalings are correction factors that normalize each gene based on its mean expression value.
Estimating the number of factors
For most datasets you want to select the number of factors based on the number of gene sets and prior knowledge as well as the granularity of the expected gene programs. However, we also provide a method to estimate the number of factors. To estimate the number of factors first, run:
from spectra import K_est as kst
L = kst.estimate_L(adata, attribute = "cell_type", highly_variable = True)
Fitting via EM
For smaller problems we can use a memory intensive EM algorithm instead
X = adata.X
A = binary adjacency matrix
model = spc.SPECTRA_EM(X = X, A= A, T = 4)
model.fit()