The goal of sfweight is to create a tidier and more streamlined interface to the spdep package. The spdep package has an idiosyncratic syntax that can be difficult to fit into a typical data science workflow. sfweight creates a simpler interface to the spdep package.
The intention behind sfweight is implement a simpler, but stricter workflow that enables the creation of neighbors, spatial weights, and spatially lagged variables. This will be accomplished by decoupling neighbors from weights and utilizing list objects.
sfweight uses sf objects whereas spdep is more flexible with the types of input objects vaialable.
You can install the development version from GitHub with
remotes::install_github("Josiahparry/sfweight")We can fit a spatial Durbin model by calculating spatially lagged predictors.
library(sfweight)
library(tidyverse)
acs_lagged <- acs %>%
mutate(nb = st_neighbors(geometry),
wts = st_weights(nb),
trans_lag = st_lag(by_pub_trans, nb, wts),
bach_lag = st_lag(bach, nb, wts))
durbin_lm <- lm(med_house_income ~ trans_lag + by_pub_trans + bach_lag + bach,
data = acs_lagged)
broom::tidy(durbin_lm)
#> # A tibble: 5 x 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 56187. 9812. 5.73 3.76e- 8
#> 2 trans_lag -13479. 28078. -0.480 6.32e- 1
#> 3 by_pub_trans -43067. 18841. -2.29 2.33e- 2
#> 4 bach_lag -40154. 28287. -1.42 1.57e- 1
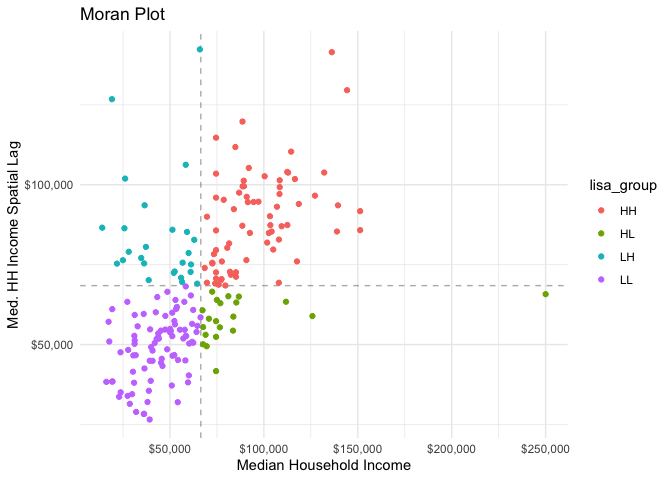
#> 5 bach 153955. 21490. 7.16 1.51e-11We can create a Moran plot by creating a spatially lagged variable.
Additionally the function categorize_lisa() will categorize high-high,
high-low, etc., groupings of these variables.
acs_lagged %>%
mutate(inc_lag = st_lag(med_house_income, nb, wts),
lisa_group = categorize_lisa(med_house_income, inc_lag)) %>%
ggplot(aes(med_house_income, inc_lag, color = lisa_group)) +
geom_vline(aes(xintercept = mean(med_house_income)), lty = 2, alpha = 1/3) +
geom_hline(aes(yintercept = mean(inc_lag)), lty = 2, alpha = 1/3) +
geom_point() +
labs(title = "Moran Plot",
y = "Med. HH Income Spatial Lag",
x = "Median Household Income") +
theme_minimal() +
scale_x_continuous(labels = scales::dollar) +
scale_y_continuous(labels = scales::dollar)We can also calculate the Local Moran’s I for each observation using the
function local_moran() this will create a dataframe column containing
the I, expected I, variance, Z-value, and P-value for each observation.
You can extract this using tidyr::unpack(). In order to do so you need
to cast as a tibble then cast back to an sf object if you want to
maintain the sf class.
acs_lisa <- acs_lagged %>%
mutate(lisa = local_moran(bach, nb, wts)) %>%
as_tibble() %>%
unpack(lisa)
acs_lisa %>%
select(last_col(4:0))
#> # A tibble: 203 x 5
#> ii e_ii var_ii z_ii p_ii
#> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0.315 -0.00495 0.138 0.863 0.194
#> 2 -0.137 -0.00495 0.328 -0.231 0.592
#> 3 1.22 -0.00495 0.245 2.47 0.00678
#> 4 -0.253 -0.00495 0.161 -0.618 0.732
#> 5 -0.0363 -0.00495 0.245 -0.0634 0.525
#> 6 1.23 -0.00495 0.245 2.49 0.00637
#> 7 0.379 -0.00495 0.138 1.03 0.150
#> 8 0.103 -0.00495 0.195 0.245 0.403
#> 9 0.0474 -0.00495 0.494 0.0745 0.470
#> 10 0.195 -0.00495 0.245 0.405 0.343
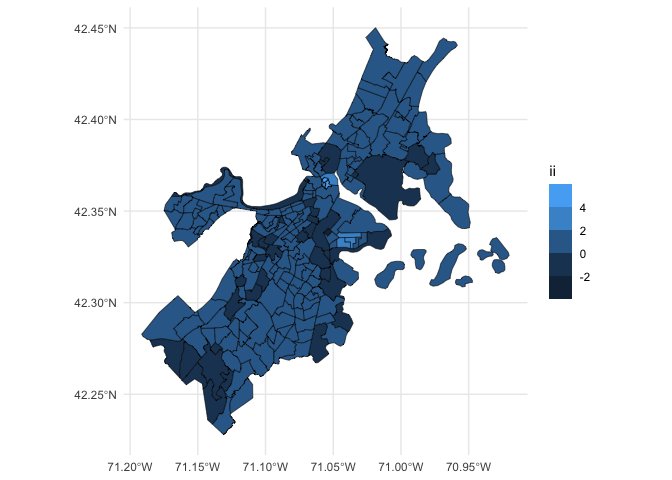
#> # … with 193 more rowslibrary(sf)
#> Linking to GEOS 3.8.1, GDAL 3.1.4, PROJ 6.3.1
acs_lisa %>%
st_as_sf() %>%
ggplot(aes(fill = ii)) +
geom_sf(color = "black", lwd = 0.2) +
scale_fill_binned(n.breaks = 5) +
theme_minimal()str(acs)
#> sf[,5] [203 × 5] (S3: sf/tbl_df/tbl/data.frame)
#> $ fips : chr [1:203] "25025092101" "25025100603" "25025010103" "25025070402" ...
#> $ med_house_income: num [1:203] 52924 86659 31218 25750 68500 ...
#> $ by_pub_trans : num [1:203] 0.3208 0.0945 0.1815 0.2229 0.199 ...
#> $ bach : num [1:203] 0.124 0.305 0.405 0.141 0.208 ...
#> $ geometry :sfc_MULTIPOLYGON of length 203; first list element: List of 1
#> ..$ :List of 1
#> .. ..$ : num [1:136, 1:2] -71.1 -71.1 -71.1 -71.1 -71.1 ...
#> ..- attr(*, "class")= chr [1:3] "XY" "MULTIPOLYGON" "sfg"
#> - attr(*, "sf_column")= chr "geometry"
#> - attr(*, "agr")= Factor w/ 3 levels "constant","aggregate",..: NA NA NA NA
#> ..- attr(*, "names")= chr [1:4] "fips" "med_house_income" "by_pub_trans" "bach"We can get neighbors based on Queen contiguities with st_neighbors().
nbs <- st_neighbors(acs)
nbs[1:5]
#> [[1]]
#> [1] 2 15 168 171 172 179 180
#>
#> [[2]]
#> [1] 1 71 180
#>
#> [[3]]
#> [1] 45 50 92 122
#>
#> [[4]]
#> [1] 30 84 127 135 136 138
#>
#> [[5]]
#> [1] 34 87 100 108If needed, we can also identify the cardinalities from the neighbors list as well.
st_cardinalties(nbs)
#> [1] 7 3 4 6 4 4 7 5 2 4 8 5 6 1 9 5 4 6 5 8 8 3 5 7 4
#> [26] 5 3 4 4 4 4 6 4 5 6 5 8 7 5 2 8 6 5 10 5 4 5 3 5 4
#> [51] 3 9 4 7 6 7 4 6 7 7 4 10 5 6 5 5 4 4 9 4 3 4 3 4 3
#> [76] 5 2 8 8 11 7 8 8 5 5 5 5 9 6 5 7 11 10 3 6 6 3 5 2 6
#> [101] 6 7 5 4 6 4 5 9 4 9 4 5 7 4 6 3 5 5 4 4 6 6 6 6 5
#> [126] 7 8 4 4 5 7 3 6 4 11 7 3 7 5 9 6 4 4 5 7 5 6 5 5 6
#> [151] 7 9 4 7 8 7 6 6 6 7 6 5 8 4 6 6 7 6 8 7 7 4 7 6 9
#> [176] 5 7 4 9 5 8 4 5 5 6 6 5 5 6 3 5 6 6 6 5 3 5 6 6 6
#> [201] 3 5 6We can get the weights from the neighbor contiguities as well. By
default, st_weights() uses row standardization.
wts <- st_weights(nbs)
wts[1:5]
#> [[1]]
#> [1] 0.1428571 0.1428571 0.1428571 0.1428571 0.1428571 0.1428571 0.1428571
#>
#> [[2]]
#> [1] 0.3333333 0.3333333 0.3333333
#>
#> [[3]]
#> [1] 0.25 0.25 0.25 0.25
#>
#> [[4]]
#> [1] 0.1666667 0.1666667 0.1666667 0.1666667 0.1666667 0.1666667
#>
#> [[5]]
#> [1] 0.25 0.25 0.25 0.25We can also calculate the spatial lag with the weights and neighbors.
inc_lag <- st_lag(acs$med_house_income, nbs, wts)
inc_lag[1:5]
#> [1] 63968.57 65019.00 59271.38 86385.17 73962.88If we have point data we can also identify the k-nearest neighbors with
st_knn(). For an example we can use the airbnb dataset that’s
imported with sfweight.
airbnb
#> Simple feature collection with 3799 features and 4 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -71.1728 ymin: 42.23576 xmax: -70.99595 ymax: 42.39549
#> Geodetic CRS: WGS 84
#> # A tibble: 3,799 x 5
#> id neighborhood room_type price geometry
#> * <dbl> <chr> <chr> <dbl> <POINT [°]>
#> 1 3781 East Boston Entire home/apt 125 (-71.02991 42.36413)
#> 2 5506 Roxbury Entire home/apt 145 (-71.09559 42.32981)
#> 3 6695 Roxbury Entire home/apt 169 (-71.09351 42.32994)
#> 4 8789 Downtown Entire home/apt 99 (-71.06265 42.35919)
#> 5 10730 Downtown Entire home/apt 150 (-71.06185 42.3584)
#> 6 10813 Back Bay Entire home/apt 179 (-71.08904 42.34961)
#> 7 10986 North End Entire home/apt 125 (-71.05075 42.36352)
#> 8 16384 Beacon Hill Private room 50 (-71.07132 42.3581)
#> 9 18711 Dorchester Entire home/apt 154 (-71.06096 42.32212)
#> 10 22195 Back Bay Private room 115 (-71.0793 42.34558)
#> # … with 3,789 more rowsairbnb_knn <- st_knn(airbnb)
airbnb_knn[1:5]
#> [[1]]
#> [1] 3091
#>
#> [[2]]
#> [1] 21
#>
#> [[3]]
#> [1] 2886
#>
#> [[4]]
#> [1] 1068
#>
#> [[5]]
#> [1] 203Point based weights implemented based on Luc Anselin and Grant Morrison’s notes.
Inverse distance band
airbnb_idw <- st_inverse_weights(airbnb$geometry, airbnb_knn)
airbnb_idw[1]
#> [[1]]
#> [1] 72.85418 94.82628 80.81517 76.98118 77.47322 207.58305 90.54686
#> [8] 140.95536 89.09559 130.49453 168.12971 76.88485 132.13504 169.34664
#> [15] 123.56357 110.04713 462.67599 73.68500 491.86866 88.92867 91.93710
#> [22] 391.89760 73.37702 81.09685 107.10884 139.86709 80.59692 111.15096
#> [29] 113.25885 126.51082 113.95462 107.27650 107.80669 108.08046 106.77599
#> [36] 98.56036 96.98179 105.01340 93.91173 91.75525 98.75033 94.91645
#> [43] 106.71431 86.72350 104.85322 80.03740 85.86828 78.68751 91.18164
#> [50] 80.82558 91.50620 87.66654 91.08201 78.62795 109.01413 94.83290
#> [57] 144.36684 133.21065 159.53591 121.62878 103.27084 108.61908 223.55088
#> [64] 132.34225 93.48938 98.53665 195.96026 272.30270 95.61728 150.25611
#> [71] 919.21712 113.75560 143.05836 135.91442 139.71490 106.91507 124.54847
#> [78] 153.71365 153.71365 153.71365 148.79092 74.75811Available kernels are:
- uniform
- triangular
- epanechnikov
- quartic
- gaussian
airbnb_gauss <- st_kernel_weight(airbnb$geometry, airbnb_knn, "gaussian")
airbnb_gauss[1]
#> [[1]]
#> [1] 2.506628 1.520893 1.866407 1.670106 1.602290 1.611395 2.357012 1.813899
#> [9] 2.193421 1.794732 2.145140 2.282160 1.600493 2.153400 2.285228 2.106956
#> [17] 2.013664 2.475767 1.538029 2.479302 1.792480 1.831595 2.463717 1.531722
#> [25] 1.674815 1.989288 2.188852 1.666432 2.022398 2.038470 2.123876 2.043607
#> [33] 1.990725 1.995231 1.997536 1.986418 1.907777 1.890761 1.970839 1.855665
#> [41] 1.829317 1.909780 1.867452 1.985884 1.761789 1.969390 1.656914 1.749397
#> [49] 1.633347 1.822058 1.670281 1.826179 1.775132 1.820787 1.632287 2.005285
#> [57] 1.866483 2.207137 2.158667 2.258595 2.095253 1.954776 2.002027 2.377080
#> [65] 2.154424 1.850619 1.907527 2.339361 2.418562 1.875498 2.228826 2.498773
#> [73] 2.042146 2.201982 2.171412 2.188205 1.987620 2.112729 2.240502 2.240502
#> [81] 2.240502 2.223651 1.559591acs %>%
transmute(nb = st_neighbors(geometry),
nb_2 = st_neighbor_lag(nb, 2),
nb_cumul_2 = st_neighbor_lag_cumul(nb, 2))
#> Simple feature collection with 203 features and 3 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -71.19125 ymin: 42.22793 xmax: -70.9201 ymax: 42.45012
#> Geodetic CRS: WGS 84
#> # A tibble: 203 x 4
#> nb nb_2 nb_cumul_2 geometry
#> * <list> <list> <list> <MULTIPOLYGON [°]>
#> 1 <int [… <int [1… <int [24]> (((-71.06249 42.29221, -71.06234 42.29273, -71.0…
#> 2 <int [… <int [6… <int [9]> (((-71.05147 42.28931, -71.05136 42.28933, -71.0…
#> 3 <int [… <int [1… <int [17]> (((-71.11093 42.35047, -71.11093 42.3505, -71.11…
#> 4 <int [… <int [1… <int [24]> (((-71.06944 42.346, -71.0691 42.34661, -71.0688…
#> 5 <int [… <int [9… <int [13]> (((-71.13397 42.25431, -71.13353 42.25476, -71.1…
#> 6 <int [… <int [1… <int [16]> (((-71.04707 42.3397, -71.04628 42.34037, -71.04…
#> 7 <int [… <int [1… <int [20]> (((-71.01324 42.38301, -71.01231 42.38371, -71.0…
#> 8 <int [… <int [8… <int [13]> (((-71.00113 42.3871, -71.001 42.38722, -71.0007…
#> 9 <int [… <int [1… <int [16]> (((-71.05079 42.32083, -71.0506 42.32076, -71.05…
#> 10 <int [… <int [1… <int [15]> (((-71.11952 42.28648, -71.11949 42.2878, -71.11…
#> # … with 193 more rows