Open Source Brain Showcase project containing examples of Neurodata Without Borders (NWB) data. OSB is developing the infrastructure to visualise and analyse experimental data in neuroscience which has been shared publicly in NWB format.
This repository will contain some example datasets which we will convert to NWB to test this functionality.
The target format will be NWB v2.0 and we intend to make use of PyNWB for reading/writing the NWB files.
The script simple.py illustrates a minimal Python script necessary to generate a valid NWB file.
To run this, install PyNWB and run:
python simple.py
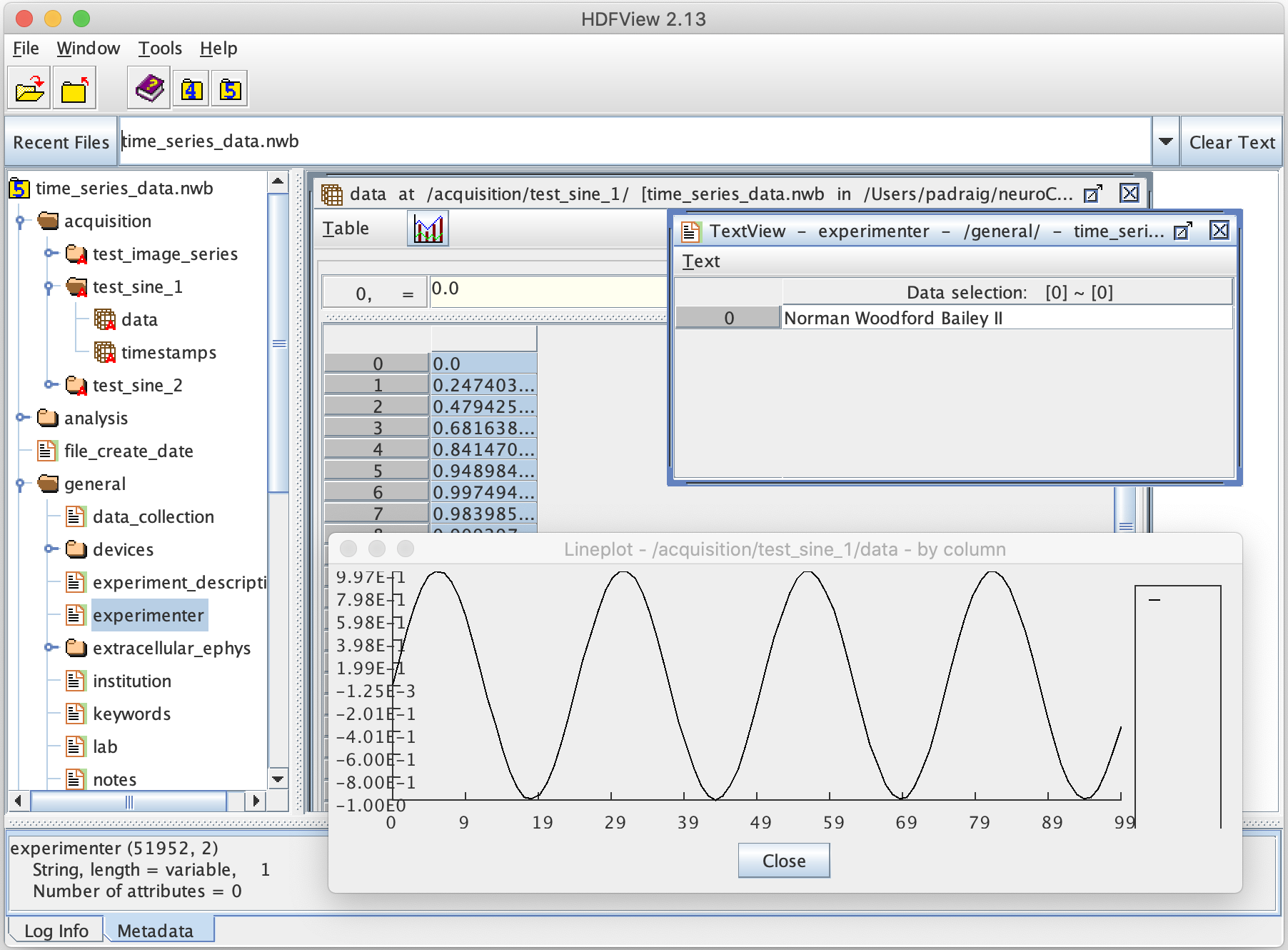
It will produce a file simple_example.nwb in HDF5 format. To view the contents of this file (or any file in HDF5) you can use HDFView. However, this NWB file doesn't have any real data in it yet. The script time_series_data.py will add some simple time series traces (sine waves) and a set of images to the NWB file to provide some example data
python time_series_data.py
The generated file can be loaded into HDFView and the structure of groups/datasets for NWB explored. Navigating to /acquisition/test_sine_1/data, opening the dataset, and then generating a Line Plot from it results in:
Other information located in the NWB file about the experiment (e.g. at /general/experimenter) can also be viewed.
However, HDFViewer doesn't treat NWB files differently from any other HDF5 file, and there are no special provisions for the types of data and metadat present in such files.
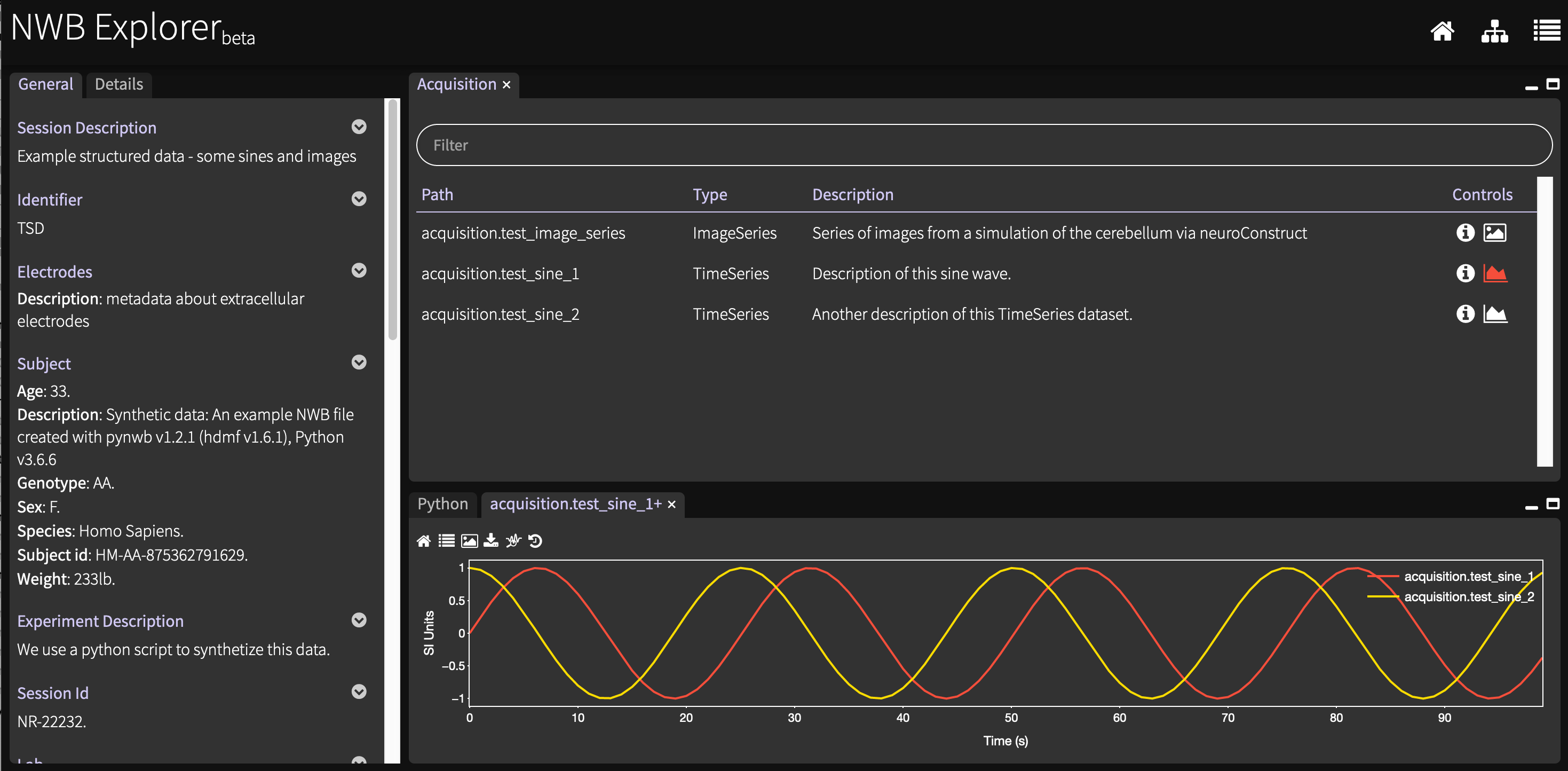
NWB Explorer (source) on the other hand does know about the structure of NWB data files, and presents this in an easily accessible way. Go to http://nwbexplorer.opensourcebrain.org and click on "Simple time series":
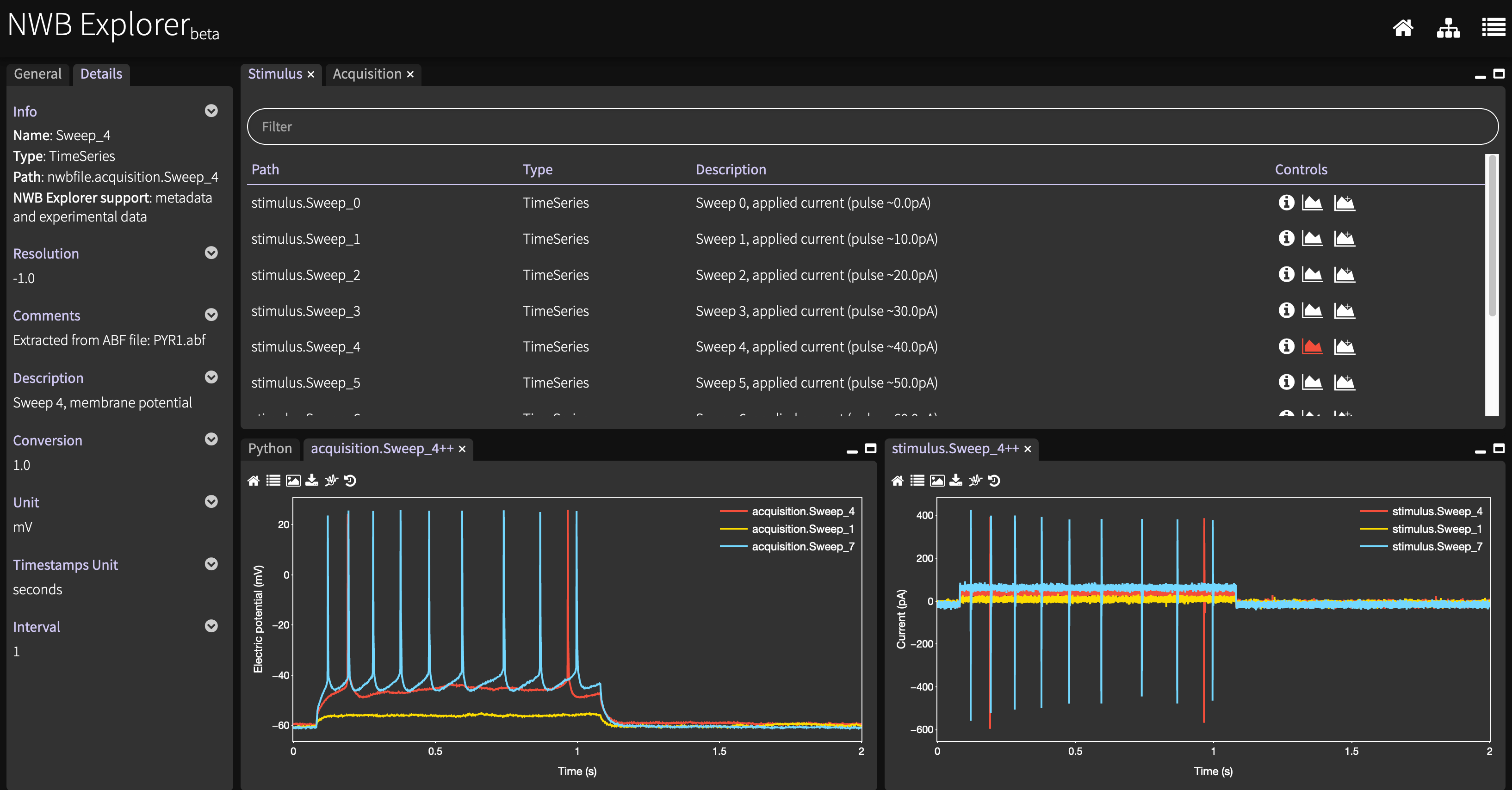
The scripts in this directory convert data which has been shared in Axon Binary Format (ABF) to NWB.
See this Jupyter notebook for the details of the conversion.
The resultant NWB files (e.g. this) can be viewed with NWB Explorer by pressing one of the links labelled Ferguson et al. 2015:
An example of converting data in MATLAB format into NWB can be found here.
This data can be viewed with NWB Explorer by pressing Lantyer et al. 2018.
An example of converting data in Igor Pro format into NWB can be found here.
This data can be viewed with NWB Explorer by pressing Lanore et al. 2019.
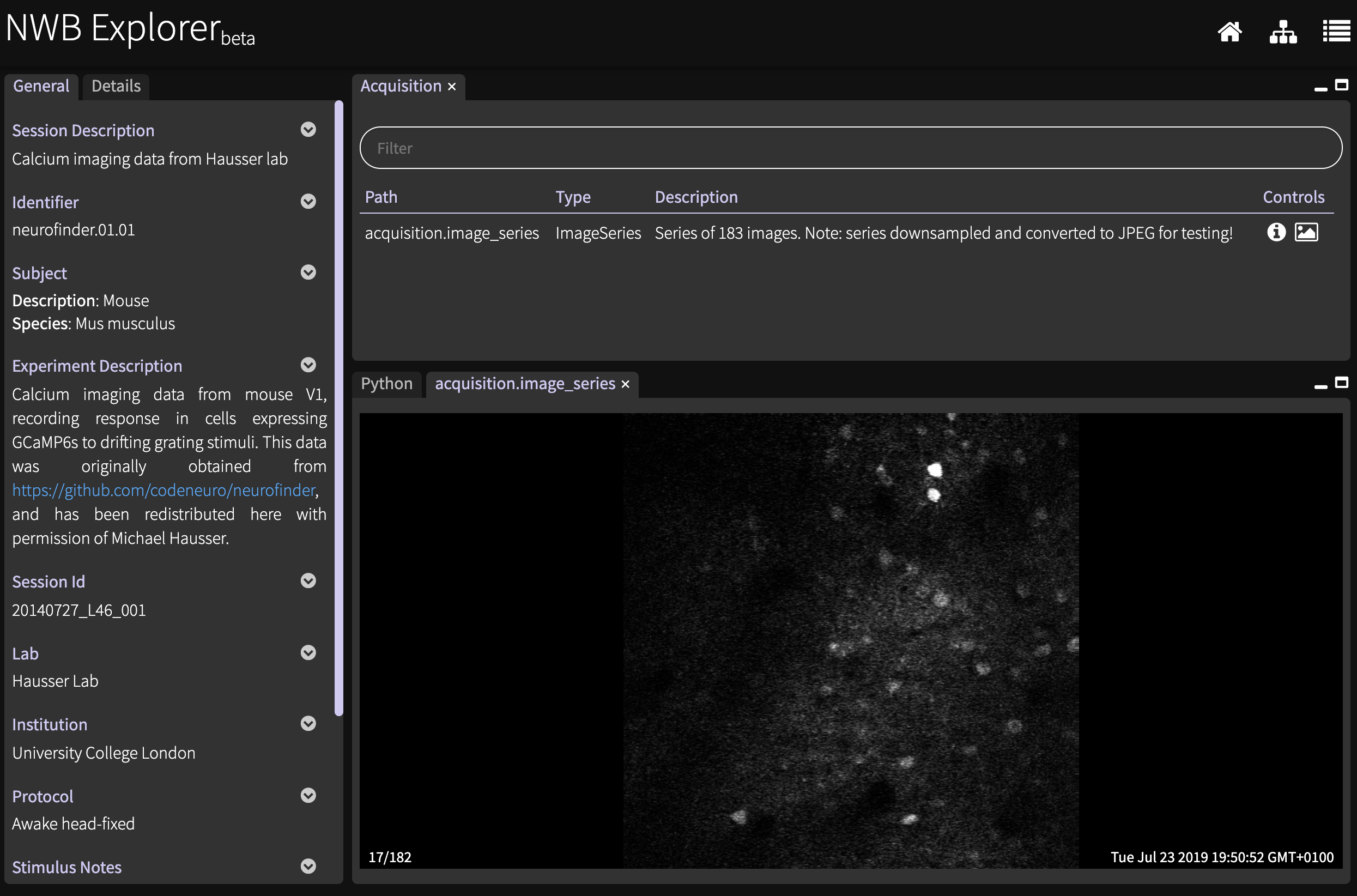
An example of converting imaging data into NWB can be found here.
This data can be viewed with NWB Explorer by pressing Packer et al. 2015.
There are a list of potentially interesting datasets here containing publicly available experimental data which could be converted to NWB format. Suggestions for more are welcome!!
Ongoing work on NWB Explorer can be followed here.
We are currently planning a new version of the Open Source Brain website (OSBv2) which will have a much deeper integration of model sharing (in NeuroML format) and data sharing (in NWB format) into a unified platform for collaborating and sharing research outputs in neuroscience.
Stay tuned!