This is the original implementation of MassFormer, a graph transformer for small molecule MS/MS prediction. Check out the preprint on arxiv.
We recommend using conda. Three conda yml files are provided in the env/ directory (cpu.yml, cu101.yml, cu102.yml), providing different pytorch installation options (CPU-only, CUDA 10.1, CUDA 10.2). They can be trivially modified to support other versions of CUDA.
To set up an environment, run the command conda env create -f ${CONDA_YAML}, where ${CONDA_YAML} is the path to the desired yaml file.
Note: this step requires a Windows System or Virtual Machine
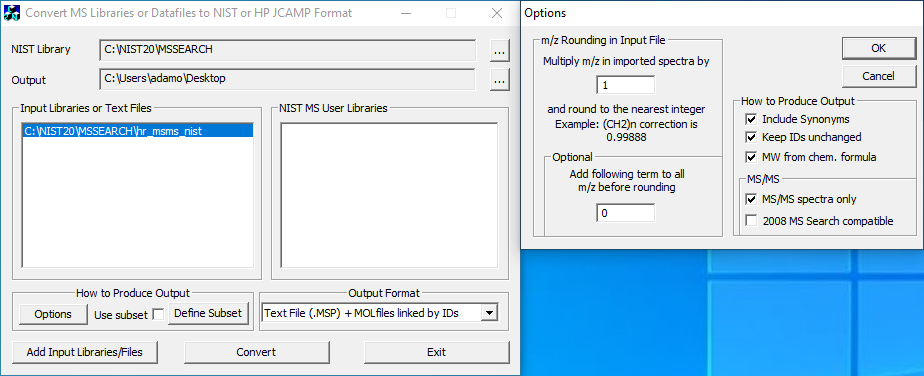
The NIST 2020 LC-MS/MS dataset can be purchased from an authorized distributor. The spectra and associated compounds can be exported to MSP/MOL format using the included lib2nist software. There is a single MSP file which contains all of the mass spectra, and multiple MOL files which include the molecular structure information for each spectrum (linked by ID). We've included a screenshot describing the lib2nist export settings.
There is a minor bug in the export software that sometimes results in errors when parsing the MOL files. To fix this bug, run the script python mol_fix.py ${MOL_DIR}, where ${MOL_DIR} is a path to the NIST export directory with MOL files.
The MassBank of North America (MB-NA) data is in MSP format, with the chemical information provided in the form of a SMILES string (as opposed to a MOL file). It can be downloaded from the MassBank website, under the tab "LS-MS/MS Spectra".
We recommend creating a directory called data/ and placing the downloaded and uncompressed data into a folder data/raw/.
To parse both of the datasets, run parse_and_export.py. Then, to prepare the data for model training, run prepare_data.py. By default the processed data will end up in data/proc/.
Our implementation uses Weights and Biases (W&B) for logging and visualization. For full functionality, you must set up a free W&B account.
A default config file is provided in "config/template.yml". This trains a MassFormer model on the NIST HCD spectra. Our experiments used systems with 32GB RAM, 1 Nvidia RTX 2080 (11GB VRAM), and 6 CPU cores.
The config/ directory has a template config file template.yml and 8 files corresponding to the experiments from the paper. The template config can be modified to train models of your choosing.
To train a template model without W&B with only CPU, run python runner.py -w False -d -1
To train a template model with W&B on CUDA device 0, run python runner.py -w True -d 0
To reproduce a model from one of the experiments in Table 2 or Table 3 from the paper, run python runner.py -w True -d 0 -c ${CONFIG_YAML} -n 5 -i ${RUN_ID}, where ${CONFIG_YAML} refers to a specific yaml file in the config/ directory and ${RUN_ID} refers to an arbitrary but unique integer ID.
The explain.py script can be used to reproduce the visualizations in the paper, but requires a trained model saved on W&B (i.e. by running a script from the previous section).
To reproduce a visualization from Figures 2,3,4,5, run python explain.py ${WANDB_RUN_ID} --wandb_mode=online, where ${WANDB_RUN_ID} is the unique W&B run id of the desired model's completed training script. The figues will be uploaded as PNG files to W&B.
The W&B sweep config files that were used to select model hyperparameters can be found in the sweeps/ directory. They can be initialized using wandb sweep ${PATH_TO_SWEEP}.