CT_PVAE is a framework that uses self-supervised learning for computed tomography reconstruction from sparse sinograms (projection measurements). The framework is probabilisitic, inferring a prior distribution from sparse sinograms on a dataset of objects and calculating the posterior for the object reconstruction on each sinogram. The associated paper is available on arXiv and was published in the NeurIPS 2022 Machine Learning and the Physical Sciences Workshop.
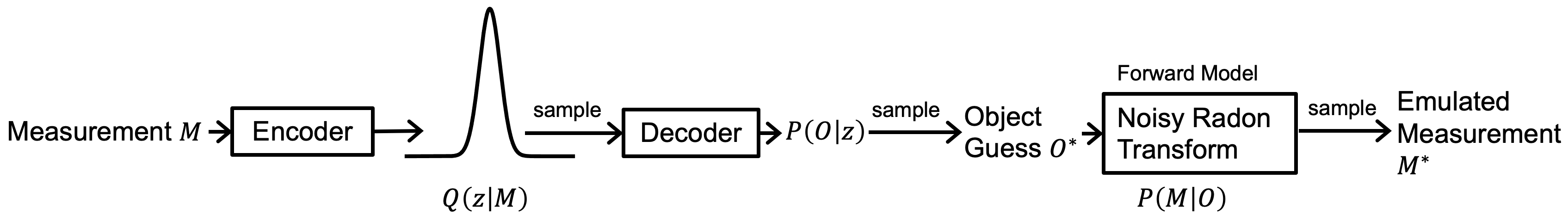
The figure below shows the overview of the end-to-end CT_PVAE pipeline.
The main algorithm comprising the CT_PVAE is inspired by the variational autoencoder. This repository allows creation of synthetic object datasets and generation of corresponding noisy, sparse sinograms. Object reconstruction from the sparse sinograms is performed with the physics-informed variational autoencoder. Code is included for visualization and comparison of results.
First, navigate to the folder where you want the repository and clone the repository:
git clone https://github.com/vganapati/CT_PVAE.git
Create a conda environment:
cd CT_PVAE
conda env create -f environment.yml
conda activate CT
Once you're done with the above step, you need to use pip install to finish installing all the dependencies, using:
pip install --upgrade -r requirements_upgrade.txt
pip install -r requirements.txt
pip install -U hpo-uq==1.3.14
And you're all set!
These are instructions to create a small synthetic dataset and quickly check that the code is working. Due to the fast training times and small size of the dataset, reconstruction results are expected to be poor. See Reproducing Paper Figures for instructions on creating larger datasets and using longer training times.
Activate your environment if not already activated:
conda activate CT
Navigate to the CT_PVAE directory. Next, set your PYTHONPATH to include the current directory:
export PYTHONPATH=$PYTHONPATH:$(pwd)
Run the following to create a synthetic foam dataset of 50 examples, saved in the subfolder dataset_foam of the current directory:
python scripts/create_foam_images.py -n 50
python scripts/images_to_sinograms.py -n 50
Finally, train the physics-informed variational autoencoder; results are saved in subfolder ct_vae_test of the current directory.
python bin/main_ct_vae.py --input_path dataset_foam --save_path ct_vae_test -b 5 --pnm 1e4 -i 1000 --td 50 --normal --visualize --nsa 20 --ns 2 --api 20 --pnm_start 1e3 --random --algorithms gridrec --train
There are several options for running the reconstruction algorithm with python bin/main_ct_vae.py.
usage: main_ct_vae.py [-h] [--ae ADAM_EPSILON] [-b BATCH_SIZE] [--ns NUM_SAMPLES] [--det] [--dp DROPOUT_PROB]
[--en EXAMPLE_NUM] [-i NUM_ITER] [--ik INTERMEDIATE_KERNEL] [--il INTERMEDIATE_LAYERS]
[--input_path INPUT_PATH] [--klaf KL_ANNEAL_FACTOR] [--klm KL_MULTIPLIER] [--ks KERNEL_SIZE]
[--lr LEARNING_RATE] [--nb NUM_BLOCKS] [--nfm NUM_FEATURE_MAPS]
[--nfmm NUM_FEATURE_MAPS_MULTIPLIER] [--norm NORM] [--normal] [--nsa NUM_SPARSE_ANGLES]
[--api ANGLES_PER_ITER] [--pnm POISSON_NOISE_MULTIPLIER] [--pnm_start PNM_START]
[--train_pnm] [-r RESTORE_NUM] [--random] [--restore] [--save_path SAVE_PATH]
[--se STRIDE_ENCODE] [--si SAVE_INTERVAL] [--td TRUNCATE_DATASET] [--train] [--ufs] [--ulc]
[--visualize] [--pixel_dist] [--real] [--no_pad] [--toy_masks]
[--algorithms ALGORITHMS [ALGORITHMS ...]] [--no_final_eval]
Get command line args
optional arguments:
-h, --help show this help message and exit
--ae ADAM_EPSILON adam_epsilon
-b BATCH_SIZE batch size
--ns NUM_SAMPLES number of times to sample VAE in training
--det no latent variable, simply maximizes log probability of output_dist
--dp DROPOUT_PROB dropout_prob, percentage of nodes that are dropped
--en EXAMPLE_NUM example index for visualization
-i NUM_ITER number of training iterations
--ik INTERMEDIATE_KERNEL
intermediate_kernel for model_encode
--il INTERMEDIATE_LAYERS
intermediate_layers for model_encode
--input_path INPUT_PATH
path to folder containing training data
--klaf KL_ANNEAL_FACTOR
multiply kl_anneal by this factor each iteration
--klm KL_MULTIPLIER multiply the kl_divergence term in the loss function by this factor
--ks KERNEL_SIZE kernel size in model_encode_I_m
--lr LEARNING_RATE learning rate
--nb NUM_BLOCKS num convolution blocks in model_encode
--nfm NUM_FEATURE_MAPS
number of features in the first block of model_encode
--nfmm NUM_FEATURE_MAPS_MULTIPLIER
multiplier of features for each block of model_encode
--norm NORM gradient clipping by norm
--normal use a normal distribution as final distribution
--nsa NUM_SPARSE_ANGLES
number of angles to image per sample (dose remains the same)
--api ANGLES_PER_ITER
number of angles to check per iteration (stochastic optimization)
--pnm POISSON_NOISE_MULTIPLIER
poisson noise multiplier, higher value means higher SNR
--pnm_start PNM_START
poisson noise multiplier starting value, anneals to pnm value
--train_pnm if True, make poisson_noise_multiplier a trainable variable
-r RESTORE_NUM checkpoint number to restore from
--random if True, randomly pick angles for masks
--restore restore from previous training
--save_path SAVE_PATH
path to save output
--se STRIDE_ENCODE convolution stride in model_encode_I_m
--si SAVE_INTERVAL save_interval for checkpoints and intermediate values
--td TRUNCATE_DATASET
truncate_dataset by this value to not load in entire dataset; overriden when restoring a
net
--train run the training loop
--ufs use the first skip connection in the unet
--ulc uses latest checkpoint, overrides -r
--visualize visualize results
--pixel_dist get distribution of each pixel in final reconstruction
--real denotes real data, does not simulate noise
--no_pad sinograms have no zero-padding
--toy_masks uses the toy masks
--algorithms ALGORITHMS [ALGORITHMS ...]
list of initial algorithms to use
--no_final_eval skips the final evaluation
Run the following once to create the datasets (both dataset_toy and dataset_foam):
Activate your environment if not already activated:
conda activate CT
Navigate to the CT_PVAE directory. Next, set your PYTHONPATH to include the current directory:
export PYTHONPATH=$PYTHONPATH:$(pwd)
Run the following to create a synthetic foam dataset of 1,000 examples, saved in the subfolder dataset_foam of the current directory:
python scripts/create_foam_images.py -n 1000
python scripts/images_to_sinograms.py -n 1000
Run the following to create a synthetic toy dataset of 1,024 examples, saved in the subfolder dataset_toy_discrete2 of the current directory:
- python scripts/create_toy_images.py -n 1024
- python scripts/images_to_sinograms.py -n 1024 --toy
Train the physics-informed variational autoencoder with the following options:
python bin/main_ct_vae.py --input_path dataset_toy_discrete2 --save_path toy_vae -b 4 --pnm 10000 -i 100000 --td 1024 --train --nsa 1 --ik 2 --il 5 --ks 2 --nb 3 --api 2 --se 1 --no_pad --ns 10 --pnm_start 1000 --si 100000 --normal --visualize --toy_masks
To create marginal posterior probability distributions for each pixel, results saved in the subfolder toy_vae of the current directory:
python bin/main_ct_vae.py --input_path dataset_toy_discrete2 --save_path toy_vae -b 4 --pnm 10000 -i 100000 --td 1024 --nsa 1 --ik 2 --il 5 --ks 2 --nb 3 --api 2 --se 1 --no_pad --ns 10 --pnm_start 1000 --si 100000 --normal --visualize --toy_masks --pixel_dist --restore --ulc --en 0
python bin/main_ct_vae.py --input_path dataset_toy_discrete2 --save_path toy_vae -b 4 --pnm 10000 -i 100000 --td 1024 --nsa 1 --ik 2 --il 5 --ks 2 --nb 3 --api 2 --se 1 --no_pad --ns 10 --pnm_start 1000 --si 100000 --normal --visualize --toy_masks --pixel_dist --restore --ulc --en 1
python bin/main_ct_vae.py --input_path dataset_toy_discrete2 --save_path toy_vae -b 4 --pnm 10000 -i 100000 --td 1024 --nsa 1 --ik 2 --il 5 --ks 2 --nb 3 --api 2 --se 1 --no_pad --ns 10 --pnm_start 1000 --si 100000 --normal --visualize --toy_masks --pixel_dist --restore --ulc --en 2
python bin/main_ct_vae.py --input_path dataset_toy_discrete2 --save_path toy_vae -b 4 --pnm 10000 -i 100000 --td 1024 --nsa 1 --ik 2 --il 5 --ks 2 --nb 3 --api 2 --se 1 --no_pad --ns 10 --pnm_start 1000 --si 100000 --normal --visualize --toy_masks --pixel_dist --restore --ulc --en 3
Train the physics-informed variational autoencoder with the following options, results saved in the subfolder foam_vae of the current directory:
python bin/main_ct_vae.py --input_path dataset_foam --save_path foam_vae -b 10 --pnm 10000 -i 100000 --td 1000 --normal --visualize --nsa 20 --ns 2 --api 20 --pnm_start 1000 --si 100000 --random --algorithms sirt tv fbp gridrec --train
If you find our work useful in your research, please cite:
@misc{mendoza2022selfsupervised,
title={A Self-Supervised Approach to Reconstruction in Sparse X-Ray Computed Tomography},
author={Rey Mendoza and Minh Nguyen and Judith Weng Zhu and Vincent Dumont and Talita Perciano and Juliane Mueller and Vidya Ganapati},
year={2022},
eprint={2211.00002},
archivePrefix={arXiv},
primaryClass={cs.CV}
}