Repository for the paper "Machine learning to predict the source of campylobacteriosis using whole genome data"
This repository allows you to attribute sources of human campylobacter from core-genome multi-locus-sequence-type (cgMLST) data. The algorithm is named aiSource after the previously most commonly used iSource. The accuracy of this prediction should be ~84% based on realistic source composition in human cases, for a more thorough investigation of performance please consult the paper. We have also added a functionality by which you can retrain the classifier with more data, or if you wish train on other labels such as countries of origin or whatever you please.
The different files in this repository do the following.
- aiSource.py - The python script allowing you to predict sources
- aiSource_classifier_object.p - A pickle object containing the XGBoost classifier trained in our paper
- test_data.tsv - a file containing test cgMLST files in a tab delimited format
- test_aiSource_out.tsv - the corresponding output file to the test data
- requirements.txt - test file containing all required python packages. Further down we explain how to set this up
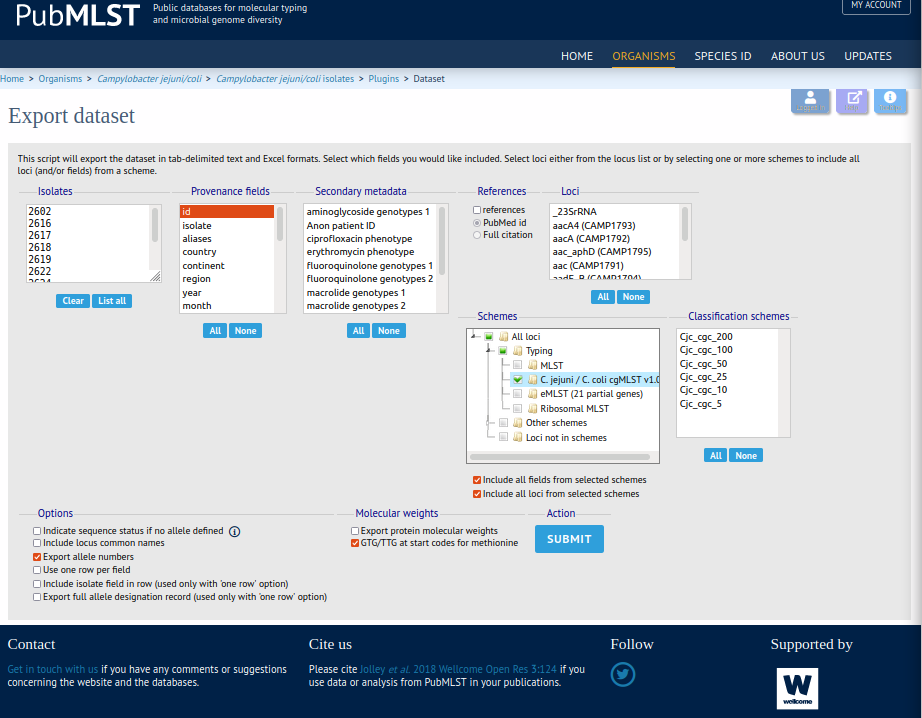
Using aiSource you can conveniently predict any human campylobacter cgMLST samples. Campylobacter sources can be downloaded from the fantastic pubmlst database, which contains a lot of different pathogens. The campylobacter database can be found here. At the bottom of the page you can export the samples using the field "Dataset" next to the Export field with the little save icon. This brings you to the Export dataset field where you can choose as many provenance fields as you want just make sure you include id and under "Schemes" you choose All loci>Typing>C. Jejuni/C. coli cgMLST v1.0 as shown below.
The resulting table should be saved with "Export table(text)" and can now be used for the prediction!
If instead you want to use the dataset we used in the paper you can find it here.
Otherwise if you have whole genomes from campy you can upload a fasta of the genome get the corresponding cgMLST patterns here. Please choose the C. jejuni. C.coli cgMLST v1.0 in the "Please select locus/scheme" menu. The resulting table should again be saved with "Export table(text)".
first you should download this repository with git clone https://github.com/narning1992/aiSource.git
then change into the folder you just downloaded with cd aiSource.
this is where you will do the analysis so your input files should also be stored in this folder.
In order to run aiSource you will need to install a few packages and make sure that you got python 3 installed. Depending on your preferred package manager you can do the following
for pip use
pip3 install -r requirements.txt
(this is the most straightforward. See here how to install pip.)
for conda use
conda create --name aiSource python=3.8
conda activate aiSource
conda install --file requirements.txt
for virtualenv
virtualenv --python=python3.8 aisource
source aisource/bin/activate
pip3 install -r requirements.txt
That should do the trick.
Now that all packages are installed you can simply predict your cgMLST files like this
python3 aiSource.py -i <path to the cgMLST file you downloaded from pubMLST> -n <prefix for your output file>
You could do a test run with the data we provided like this:
python3 aiSource.py -i test_data.tsv -n test
The output file will be created in the same folder under <prefix>_aiSource_out.tsv. Please be aware that rows with more than 10% missingness will be removed and won't show up in the output.
The output has 7 columns. The "id" columns contains the identifier specified in the input file under id. The "predicted_source" column contains the source predicted by aiSource. The following columns contain the predicted probabilities across all sources which sum up to 1.
We have also added a functionality to retrain the classifier with the same hyperparameter. You can now either feed in more samples for source attribution or other include more sources or predict a feature of your choosing. It's really simple. You will have to divide your data into two seperate tables for training and prediction. Of course you can also use the same file for training and prediction but there is little value in predicting what has been seen in training.
For training the classifier make sure you have a column in your training data that has the desired labels. As an example we could use the "country" column in the test_data.tsv. Make sure there are enough samples for every category within the label. The classifier will remove all labels with less than 15 members. You would then retrain the classifier like this
python aiSource.py -rt country -i test_data.tsv -n country
The classifier is then retrained on 75% so we can use the residual 25% of samples for testing the accuracy which will be printed to the screen but also saved under <prefix>_retraining_accurary_report.txt. Subsequently all of the training data will be used to train the classifier and the classifier object will be stored under <prefix>_retrained_clf.p. We can then use this classifier to predict more data. In the case of our country classifier we would do it like so:
python aiSource.py -clf country_retrained_clf.p -i test_data.tsv -n country
This example is quite useless as we are predicting the data we have already seen in training. However when used you should put in new cgMLST data and it will get predicted. Technically you could predict any label you can imagine from the cgMLST data an easily adapt this for other bugs than campylobacter. We hope this will be very useful to the community