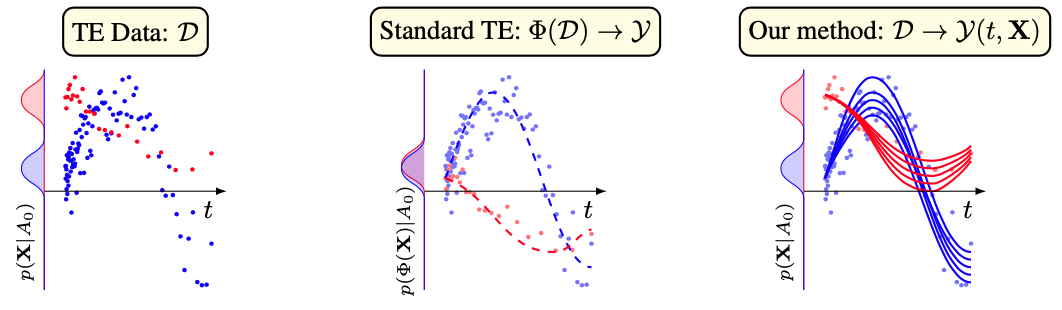
This repo holds the code, and log files for ODE Discovery for Longitudinal Heterogeneous Treatment Effects Inference by Samuel Holt, Jeroen Berrevoets, Krzysztof Kacprzyk, Zhaozhi Qian, and Mihaela van der Schaar.
We introduce a different type of solution for the Longitudinal Heterogeneous Treatment Effects Inference: a closed-form ordinary differential equation (ODE). While we still rely on continuous optimization to learn an ODE, the resulting inference machine is no longer a neural network. Doing so yields several advantages such as interpretability, irregular sampling, and a different set of identification assumptions. Above all, we consider the introduction of a completely new type of solution to be our most important contribution as it may spark entirely new innovations in treatment effects in general. In the paper we contribute a framework that can transform any ODE discovery method into a treatment effects method. Using our framework, we build an example method called Individualized Nonlinear Sparse Identification Treatment Effect (INSITE). This is tested in accepted benchmark settings used throughout the literature.
To get started:
- Clone this repo
git clone https://github.com/samholt/ODE-Discovery-for-Longitudinal-Heterogeneous-Treatment-Effects-Inference && cd ./ODE-Discovery-for-Longitudinal-Heterogeneous-Treatment-Effects-Inference- Follow the installation instructions in
setup/install.shto install the required packages.
./setup/install.shOpen up a separate terminal and run mlflow server --port=5000 to start the mlflow server.
In the main terminal, perform the following steps:
- Modify the configuration files in folder
config. The main config file that specifies baselines, datasets and other run parameters is inconfig/config.yaml - Run
python run.pyto run all baselines on all datasets. This will generate a log file in thelogsfolder. - Once a run has completed, process the log file generated output into the
logsfolder, with the scriptprocess_result_file.py. Note, you will need to edit theprocess_result_file.pyto read this generated log file, i.e., specify the path variable of where it is. This will generate the main tables as presented in the paper.
If you use our work in your research, please cite:
@inproceedings{
holt2024ode,
title={{ODE} Discovery for Longitudinal Heterogeneous Treatment Effects Inference},
author={Samuel Holt and Jeroen Berrevoets and Krzysztof Kacprzyk and Zhaozhi Qian and Mihaela van der Schaar},
booktitle={The Twelfth International Conference on Learning Representations},
year={2024},
url={https://openreview.net/forum?id=pxI5IPeWgW}
}