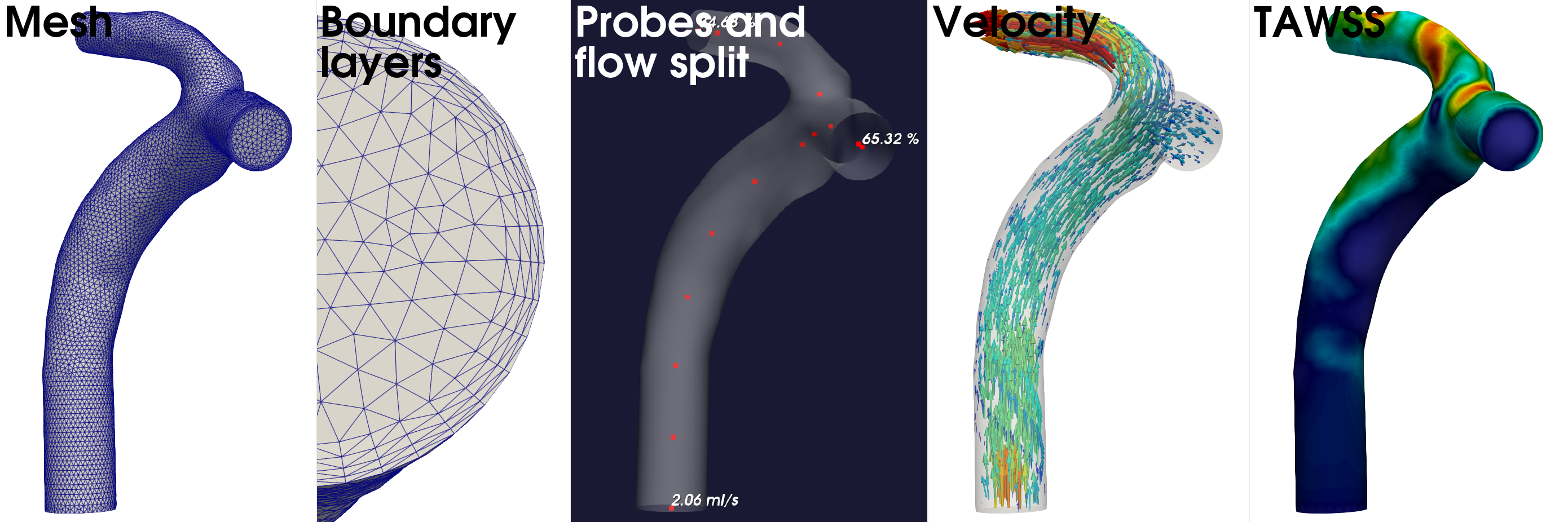
Pre-processed and simulated artery model. From left to right: A variable density volumetric mesh, zoomed in view of an outlet showing the four boundary layers, corresponding inlet flow rate, outlet flow split, and probes for velocity and pressure sampling. From the simulation results, we have shown the velocity field represented by vectors, and the time averaged wall shear stress (TAWSS) as one of the many hemodynamic indices computed by the post-processing scripts.
The Vascular Modeling Pypeline (VaMPy) is a collection of fully automated scripts used to prepare, run, and analyze cardiac and atrial morphologies. This includes pre-processing scripts for meshing and probe sampling, two Oasis problem files for simulating flow in the internal carotid artery and the left atrium, and a variety of post-processing scripts for computing WSS-based metrics, more advanced turbulence metrics, and a variety of morphological parameters in patient-specific geometries.
VaMPy has been developed by
- Aslak Wigdahl Bergersen
- Christophe Chnafa
- Henrik A. Kjeldsberg
VaMPy is licensed under the GNU GPL, version 3 or (at your option) any later version.
VaMPy is Copyright (2018-2022) by the authors.
For detailed installation notes and an introduction to VaMPy, please refer to the documentation.
For reference, VaMPy requires the following dependencies: morphMan, FEniCS, and Paramiko.
If you are on Windows, macOS or Linux you can install all the general dependencies through Anaconda.
First install Anaconda or Miniconda on your computer, and add the conda-forge channel with:
conda config --add channels conda-forge
conda config --set channel_priority strict
Then create a conda environment for morphMan and FEniCS by executing the following command in a terminal
conda create -n your_environment morphman fenics paramiko
Next, you need to install Oasis. You can do so with the following commands:
git clone https://github.com/mikaem/Oasis
pip install cppimport
pip install --editable Oasis
Finally, you are ready to clone and use the Vascular Modeling Pypeline:
git clone https://github.com/KVSLab/VaMPy.git
cd VaMPy
Please report bugs and other issues through the issue tracker at: