A 3D multi-modal medical image segmentation library in PyTorch
We strongly believe in open and reproducible deep learning research. Our goal is to implement an open-source medical image segmentation library of state of the art 3D deep neural networks in PyTorch. We also implemented a bunch of data loaders of the most common medical image datasets. The first stable release of our repository is almost ready. This project started as an MSc Thesis and is currently under further development.
Although this work was initially focused on 3D multi-modal brain MRI segmentation we are slowly adding more architectures and data-loaders. Stay tuned! More updates are coming...
Quick Start
- If you want to quickly understand the fundamental concepts, we strongly advice to check our blog post, which provides a high level overview of all the aspects of medical image segmentation and deep learning.
- Alternatively, you can create a virtual environment and install the requirements. Check the installation folder for more instructions.
- You can also take a quick glance at the manual.
- If you do not have a capable environment or device to run this projects then you could give Google Colab a try. It allows you to run the project using a GPU device, free of charge. You may try our Colab demo using this notebook:
Implemented architectures
-
U-Net3D Learning Dense Volumetric Segmentation from Sparse Annotation Learning Dense Volumetric Segmentation from Sparse Annotation
-
V-net Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation
-
ResNet3D-VAE 3D MRI brain tumor segmentation using auto-encoder regularization
-
U-Net Convolutional Networks for Biomedical Image Segmentation
-
SkipDesneNet3D 3D Densely Convolutional Networks for Volumetric Segmentation
-
HyperDense-Net A hyper-densely connected CNN for multi-modal image segmentation
-
multi-stream Densenet3D A hyper-densely connected CNN for multi-modal image segmentation
-
DenseVoxelNet Automatic 3D Cardiovascular MR Segmentation with Densely-Connected Volumetric ConvNets
-
MED3D Transfer learning for 3D medical image analysis
-
HighResNet3D On the Compactness, Efficiency, and Representation of 3D Convolutional Networks: Brain Parcellation as a Pretext Task
-
COVID-Net A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest Radiography Images
Implemented medical imaging data-loaders
-
Iseg 2017
-
Mrbrains 2018
-
MICCAI BraTs2018
-
IXI brain development Dataset
-
MICCAI Gleason 2019 Challenge
-
COVID-CT dataset
-
COVIDx dataset
-
COVID-19 CT Lung and Infection Segmentation Dataset
Preliminary results
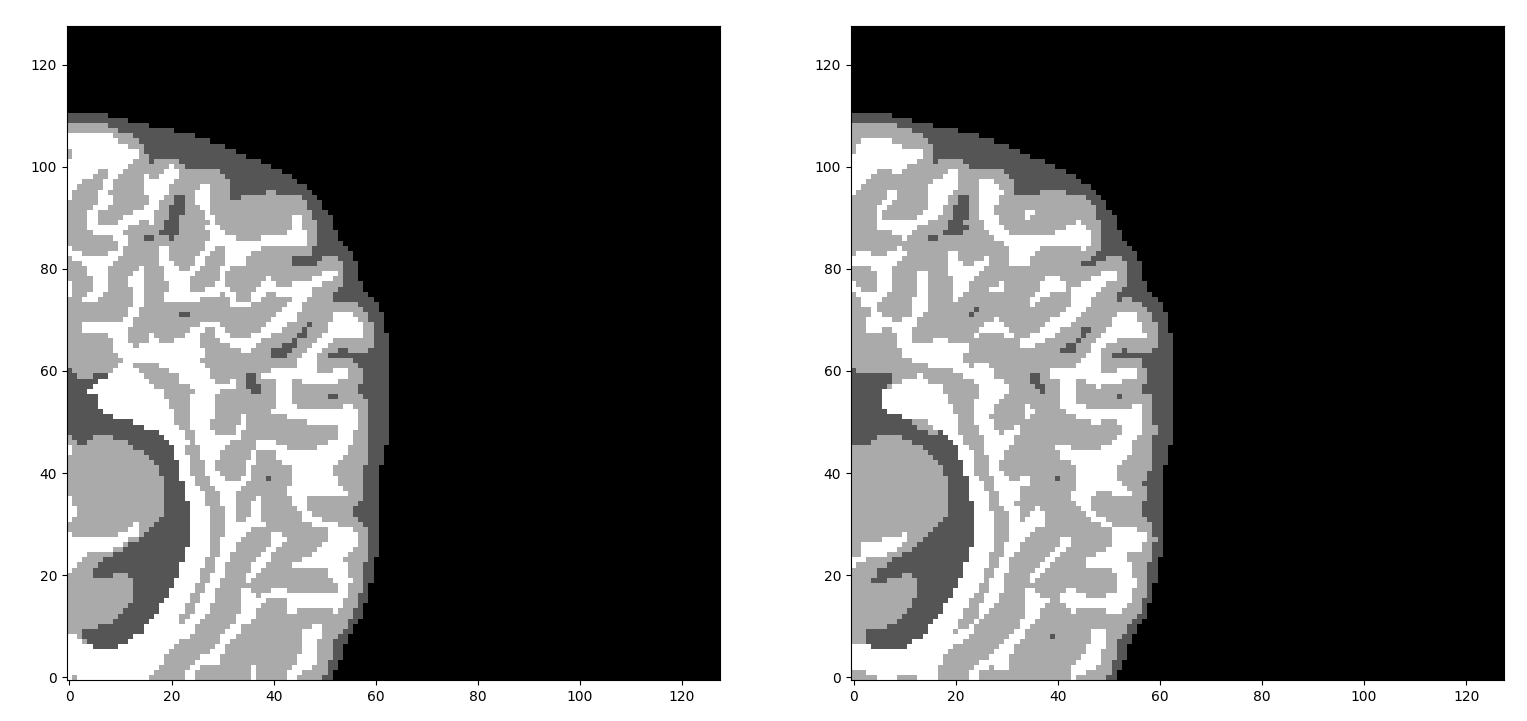
Visual results on Iseg-2017
Iseg and Mr-brains
| Model | # Params (M) | MACS(G) | Iseg 2017 DSC (%) | Mr-brains 4 classes DSC (%) |
|---|---|---|---|---|
| Unet3D | 17 M | 0.9 | 93.84 | 88.61 |
| Vnet | 45 M | 12 | 87.21 | 84.09 |
| DenseNet3D | 3 M | 5.1 | 81.65 | 79.85 |
| SkipDenseNet3D | 1.5 M | 31 | - | - |
| DenseVoxelNet | 1.8 M | 8 | - | - |
| HyperDenseNet | 10.4 M | 5.8 | - | - |
Usage
How to train your model
- For Iseg-2017 :
python ./tests/train_iseg.py --args
- For MR brains 2018 (4 classes)
python ./tests/train_mrbrains_4_classes.py --args
- For MR brains 2018 (8 classes)
python ./tests/train_mrbrains_8_classes.py --args
- For MICCAI 2019 Gleason Challenge
python ./tests/test_miccai_2019.py --args
- The arguments that you can modify are extensively listed in the manual.
New released cool features (04/2020)
- On the fly 3D total volume visualization
- Tensorboard and PyTorch 1.4 support to track training progress
- Code cleanup and packages creation
- Offline sub-volume generation
- Add Hyperdensenet, 3DResnet-VAE, DenseVoxelNet
- Fix mrbrains,Brats2018, IXI,MICCAI 2019 gleason challenge dataloaders
- Add confusion matrix support for understanding training dynamics
- Visualizations
Top priorities
- Unify/Generalize Train and Test functions
- Minimal test prediction example with pre-trained models
- Save produced 3d-total-segmentation as nifty files
Support
If you really like this repository and find it useful, please consider (★) starring it, so that it can reach a broader audience of like-minded people. It would be highly appreciated :) !
Contributing to Medical ZOO
If you find a bug, create a GitHub issue, or even better, submit a pull request. Similarly, if you have questions, simply post them as GitHub issues. More info on the contribute directory.
Current team
Ilias Papastatis
Nikolas Adaloglou
License and citation
Advice the LICENSE.md file. For usage of third party libraries and repositories please advise the respective distributed terms. It would be nice to cite the original models and datasets. If you want, you can also cite this work as:
@MastersThesis{adaloglou2019MRIsegmentation,
author = {Adaloglou Nikolaos},
title={Deep learning in medical image analysis: a comparative analysis of
multi-modal brain-MRI segmentation with 3D deep neural networks},
school = {University of Patras},
note="\url{https://github.com/black0017/MedicalZooPytorch}",
year = {2019},
organization={Nemertes}}