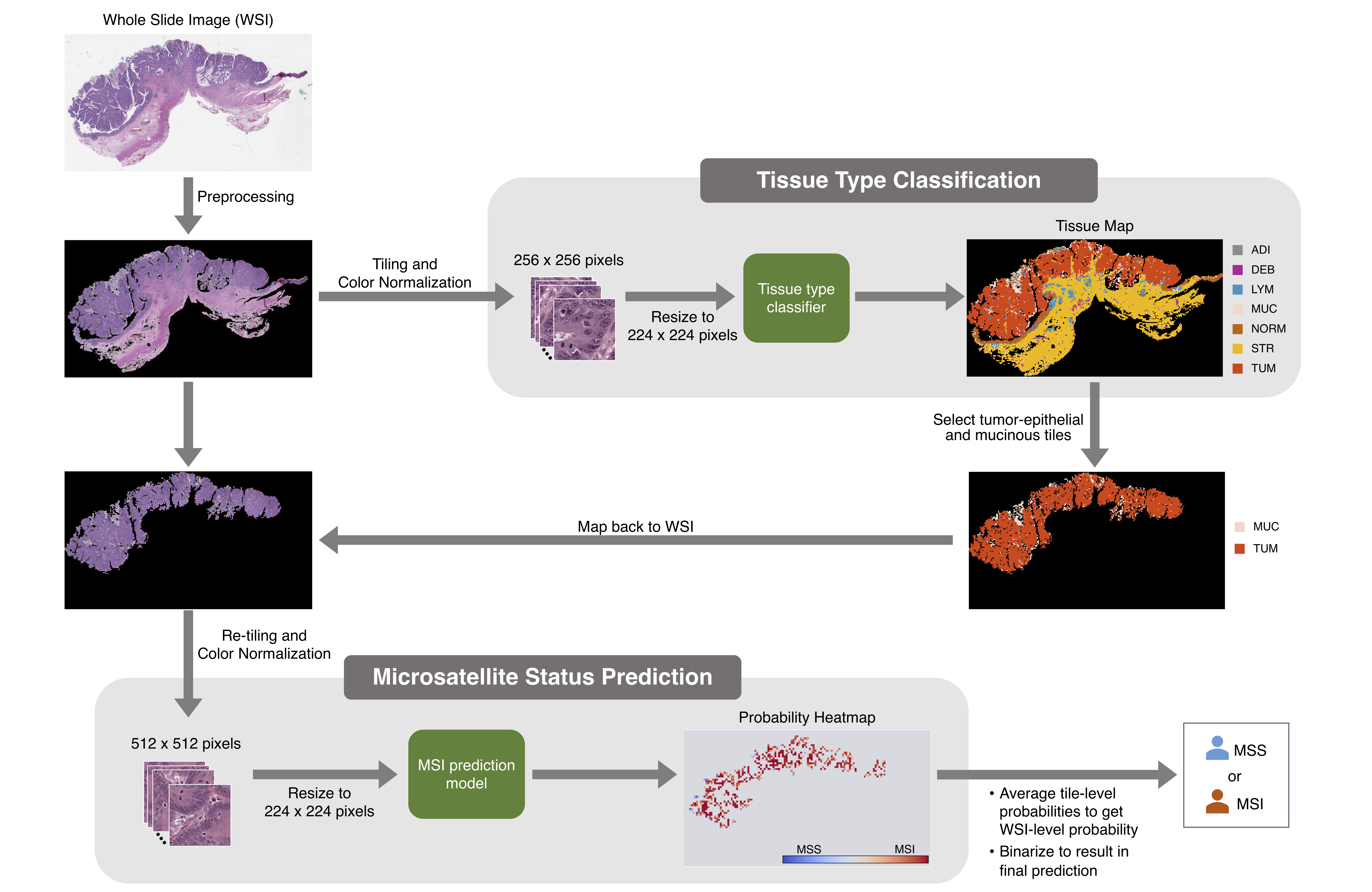
MSINet: Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study
This repository contains the code for predicting microsatellite status in colorectal cancer from H&E-stained FFPE histopathology slides.
Software Requirements
This code was developed and tested in the following settings.
OS
- Ubuntu 18.04
GPU
- Nvidia GeForce RTX 2080 Ti
Dependencies
- python (3.6.10)
- numpy: (1.18.1)
- pandas (0.25.3)
- pillow (7.0.0)
- matplotlib (3.1.0)
- scikit-learn (0.21.3)
- scikit-image (0.15.0)
- opencv-python (4.1.2.30)
- openslide-python (1.1.1)
- staintools (2.1.2)
- h5py (2.9.0)
- pytables (3.5.1)
- pytorch (1.4.0)
- torchvision (0.5.0)
- fastai (1.0.55)
Installation
-
Install Miniconda on your machine (download the distribution that comes with python3).
-
After setting up Miniconda, install OpenSlide (3.4.1):
apt-get install openslide-tools
- Create a conda environment with environment.yml:
conda env create -f environment.yml
- Activate the environment:
conda activate msinet
Demo
data collection
- Download diagnostic whole-slide images from TCGA-COAD project and TCGA-READ project using GDC Data Transfer Tool Client.
gdc-client download -m gdc_manifest_tcga_coadread.txt
- Download NCT-CRC-HE-100K and CRC-VAL-HE-7K datasets (generated by Kather et al.) from here to train and test tissue type classifier.
modify NCT-CRC-HE-100K and CRC-VAL-HE-7K datasets and train tissue type classfier
python tissue_type_classifier.py
prepare dataset for tissue type inference
python svs2tile.py
python svs_tile2hdf.py
apply tissue type classifier to generate tissue maps
python tissue_type_inference.py
prepare datasets (Stanford-CRC and TCGA-CRC) for MSI prediction
python tmap2tile.py
python tmap_tile2hdf.py
train MSI predictor on Stanford-CRC
python msi_predictor.py
evaluate MSI predictor on TCGA-CRC
python msi_inference.py
Note: please edit paths in each .py file.
Citation
Lancet Oncology 2021;22(1):132–41
@ARTICLE{Yamashita2021deep,
title = "Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study",
author = "Yamashita, Rikiya and Long, Jin and Longacre, Teri and Peng, Lan and Berry, Gerald and Martin, Brock and Higgins, John and Rubin, Daniel L and Shen, Jeanne",
journal = "Lancet Oncol.",
volume = 22,
number = 1,
pages = "132--141",
month = jan,
year = 2021,
language = "en"
}