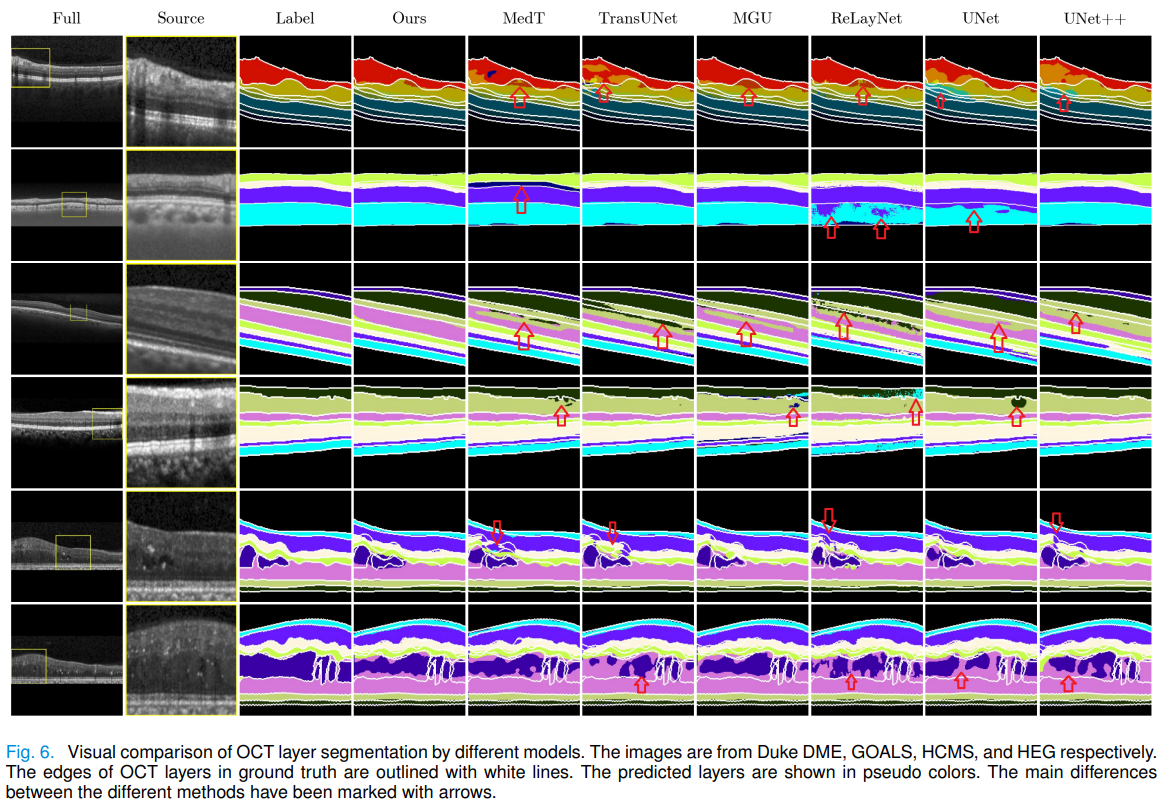
- MICCAI2022 Challenge: Glaucoma Oct Analysis and Layer Segmentation (GOALS)
- GOALS Challenge:https://aistudio.baidu.com/aistudio/competition/detail/230/0/introduction
- Implementation by PaddlePaddle/PyTorch
- Pytorch implementation of the paper "Retinal Layer Segmentation in OCT images with Boundary Regression and Feature Polarization" (accepted by TMI2023).
The project's code is constantly being updated!
- Python 3.8
- Paddle 2.3.2
- Pytorch 1.13.0
Model:TCCT/PyTorch
Project for task1:Segmentation
├── data (code for datasets)
├── tran.py (some python imports)
├── octnpy.py (parent class for OCT datasets)
├── octgen.py (child class for OCT datasets)
└── ...
├── kite (package for segmentation with torch)
├── loop_seg.py (child class for training)
├── loopback.py (parent class for training)
├── main.py
└── ...
├── nets (related models)
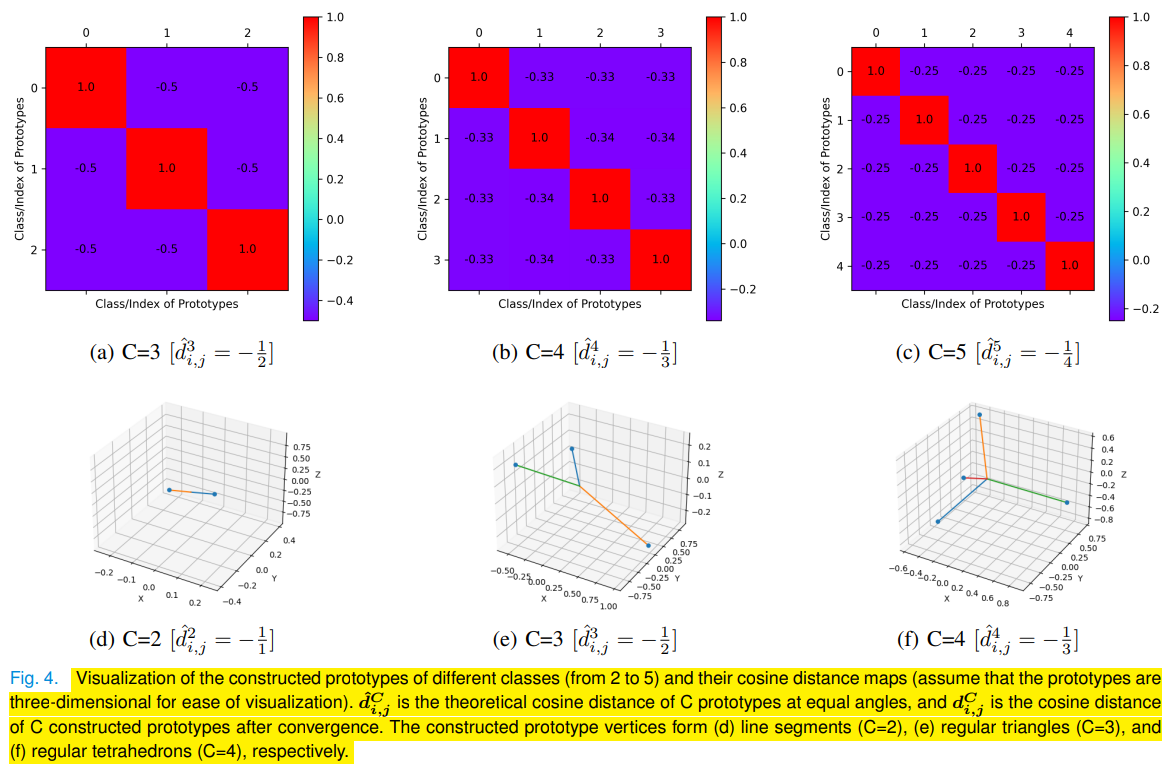
├── fcp.py (Feature Polarization Loss - file1)
├── fcs.py (Feature Polarization Loss - file2)
├── reg.py (loss functions [feature polarization & boundary regression])
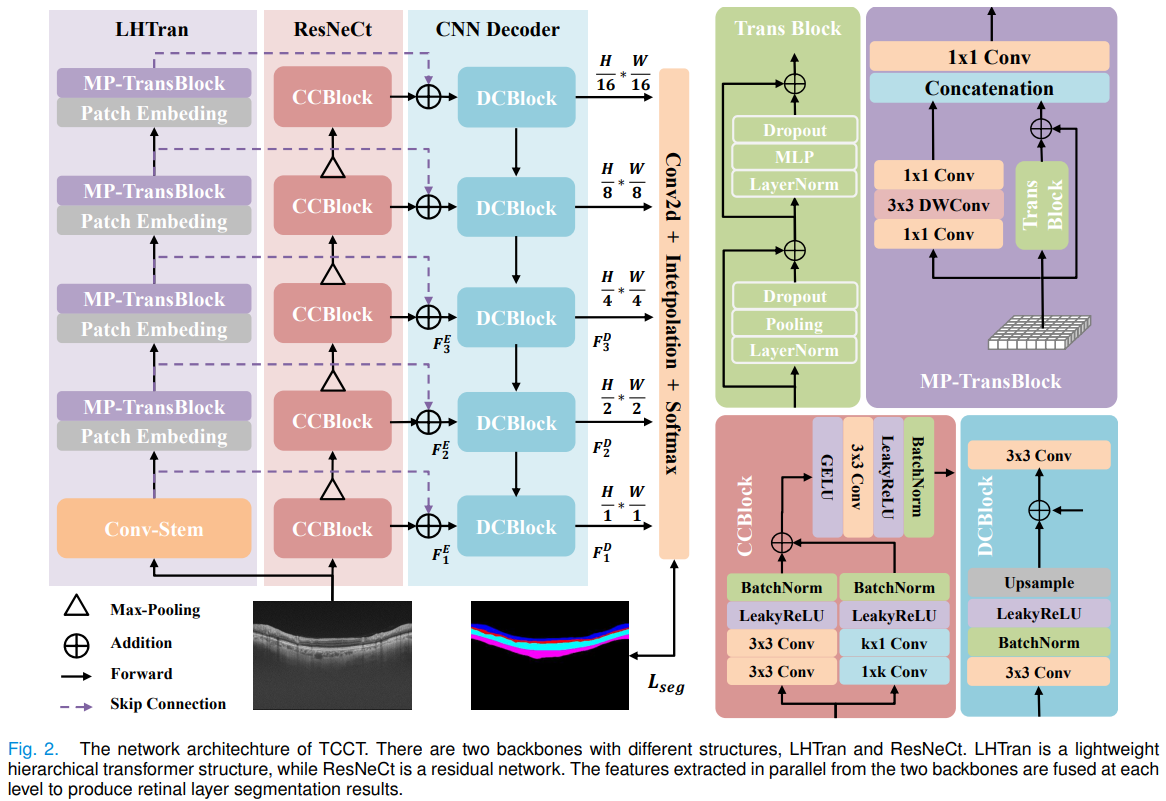
├── tcct.py (Tightly combined Cross-Convolution and Transformer)
└── ...
├── pnnx (trained weights)
├── onnx.py (code to inference OCT images with *.onnx files)
└── ...
And for the training on GOALS dataset, run the command
CUDA_VISIBLE_DEVICES=0 python kite/main.py --bs=8 --net=stc_tt --los=di --epochs=100 --db=goalsAnd for the training on HCMS dataset, run the command
CUDA_VISIBLE_DEVICES=1 python kite/main.py --bs=8 --net=stc_tt --los=di --epochs=100 --db=hcmsIf you would like to use the code, please cite our work.
Y. Tan et al., "Retinal Layer Segmentation in OCT images with Boundary Regression and Feature Polarization," in IEEE Transactions on Medical Imaging, doi: 10.1109/TMI.2023.3317072.
@article{tan2023tcct,
author={Tan, Yubo and Shen, Wen-Da and Wu, Ming-Yuan and Liu, Gui-Na and Zhao, Shi-Xuan and Chen, Yang and Yang, Kai-Fu and Li, Yong-Jie},
journal={IEEE Transactions on Medical Imaging},
title={Retinal Layer Segmentation in OCT images with Boundary Regression and Feature Polarization},
year={2023},
ISSN={1558-254X},
doi={10.1109/TMI.2023.3317072},
publisher={IEEE}
}
Model:ResNet/Paddle
- Training:
"python t2_train.py --gpu=0" - Ensemble:
"python t2_ensemble.py --root=xxx --gpu=0"