Author: Thibault Tubiana, PhD
Please read before using this script.
This script is made to facilitate the preparation and production of protein and protein/ligand, MD.
It follows the procedure described for teaching I made at the University of Bergen. You can find lectures content on this page http://tubiana.me/teaching/kjem220-molecular-modelling/ or the pdf describing all the steps on this script here: http://tubiana.me/teaching_files/biocat2020/Tutorial_Gromacs-2019.pdf
Fundamental analysis is also generated with gromacs tools (temperature/pressure/rmsd/rmsf/...), and the production trajectories are also cleaned with trjconv (imaging/protein centred/water stripped), but all the original trajectories are kept.
Feel free to make other analysis of course, like trajectory clustering with TTClust https://github.com/tubiana/TTClust 😇
- Each system is unique. This protocol and MD parameters is not adapted for all systems. If your system crash, you may have to tweak MDP parameters.
- Ligand parametrisation is "quick and dirty", For a more stable MD system you may have to tweak ACPYPE parameters (and check the hydrogens that are added with babel).
- this script only works on Linux (maybe Mac) and use the BASH syntax.
- For ligand parametrisation, I use ACPYPE (https://github.com/alanwilter/acpype) which can generate parameters for Amber, Gromacs and Charmm. Please cite this paper if you use ACPYPE: https://doi.org/10.1016/j.softx.2019.100241.
- To install ACPYPE, I sugg0est you to install first Miniconda (if you don't already have conda https://docs.conda.io/en/latest/miniconda.html) and the create a new conda environment with the command
conda create -n acpype -c conda-forge acpype - then activate the environment with
conda activate acpype - Hydrogens on ligand: openbabel. You can install it with
conda install -c conda-forge openbabel
- To install ACPYPE, I sugg0est you to install first Miniconda (if you don't already have conda https://docs.conda.io/en/latest/miniconda.html) and the create a new conda environment with the command
Here's a unique command line to create a environment with every depencencies
conda create -n gmx -c conda-forge -c salilab acpype dssp
you can activate the environment with conda activate gmx
- Make sure you have all the dependencies
- If you have a protein-ligand system, make sure acpype is installed (see parameters)
- Gromacs
- (optional) DSSP version 3
- Clone this repository with the command
git clone https://github.com/tubiana/protocolGromacs.git - Put your PDB in the repository
- Make the change you need in runGromacs.sh (See parameters)
- run the script with
bash runGromacs.sh
You have to make some changes in the script file (runGromacs).
- FILE: PDB filename without the extension (2h4g.pdb --> FILE=2h4g)
- LIGNAME: 3 letter ligand name (it has to be the same in the PDB). NOTE: The ligand name will be change to
LIGafterward. - BOXSIZE: Periodic box size in nm (between protein and box facet) default is 1.2
- BOXTYPE: Box type. Default is cubic (see http://manual.gromacs.org/documentation/5.1.4/onlinehelp/gmx-editconf.html for more details)
- NT: Number of CPU cores. Default is 8
- WATER: Water-type. Default is tip3p
- NUMBEROFREPLICAS: Number of replicas (the same simulation will be done 3 times from the minimisation). Default is 3
- FF: Force field, default is amber99sb-ildn
- SIMULATIONTIME: Simulation time in
ns. Default is 100. The script will automatically calculate and modify the number of steep according to the timestep in mdp/md_prod.mpd.
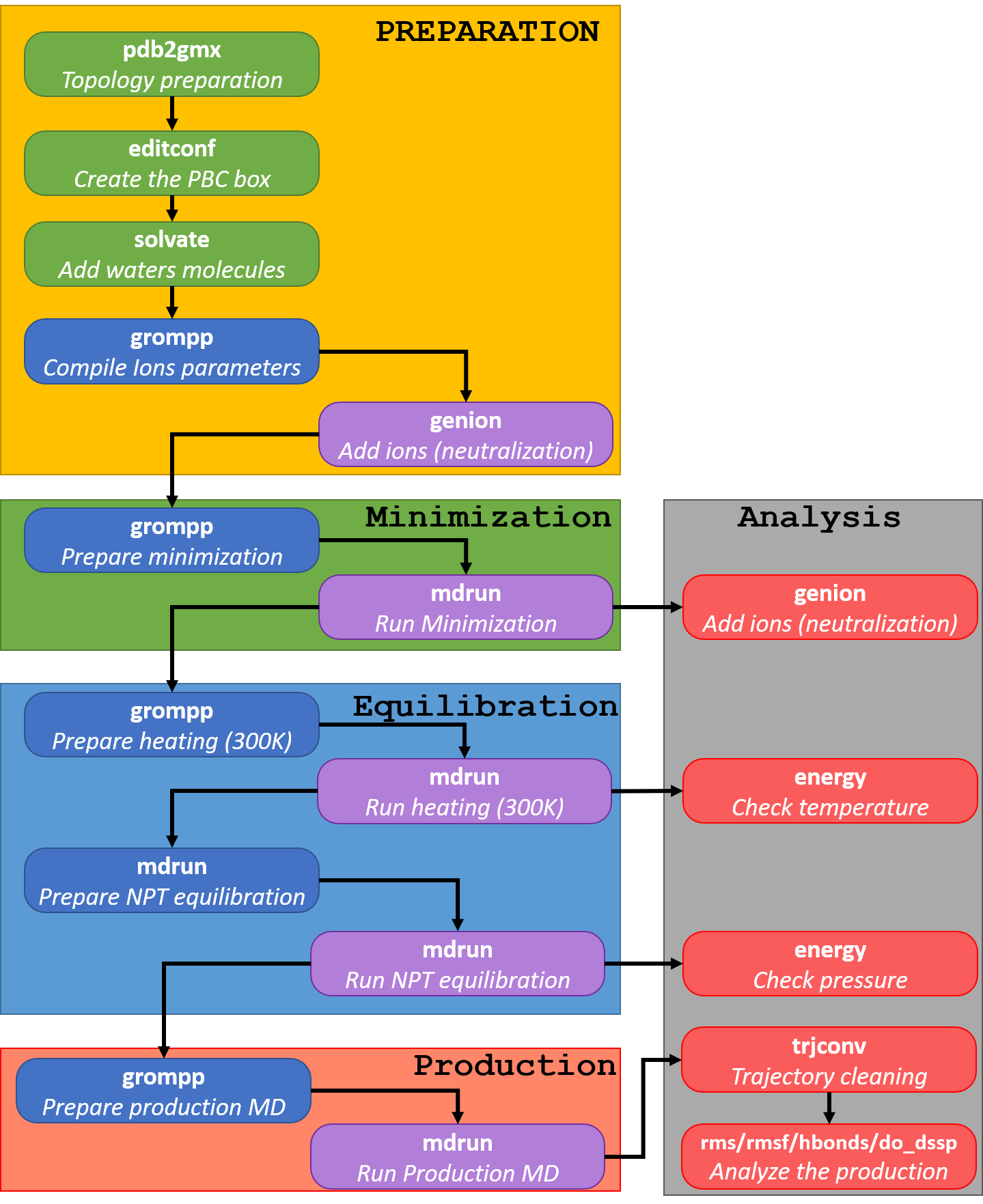
Here's a picture describing the workflow in this script, but you can find more information on each step on my tutorial http://tubiana.me/teaching_files/biocat2020/Tutorial_Gromacs-2019.pdf. You can, of course, modify my script as you want :-)
Here's a description of the folder structure after a simulation job:
|-- . #--> repo folder, the script, the initial structure and topologie files
|-- param #--> only if ligand is present, will contain receptor and ligand parameters
|-- receptor #--> receptor structure and topology
|-- ligand #--> receptor topology
|-- ligand.acpype #--> ligand topology
|-- mdp #--> original mdp parameters
|-- replica_X #--> simulation for replica number X (if 3 replica, then 3 folders)
|-- graph #--> All the output graph are saved here (rmsd,rmsf,energy.....)
|-- gro #--> Some output structures from MD are saved here
|-- mdp #--> copy of previous mdp folder
|-- results #--> contains the MD
|-- mini #--> minimisation MD files
|-- nvt #--> heationg MD files
|-- npt #--> equilibration MD files
|-- prod #--> production MD files
Have fun with MD and send me a mail if or open an issue if you have any problems, or just if you used this script and want to thanks me, I will be please to know that it was useful for someone 🙂
Thibault Tubiana.