Starfish
Introduction
The goal of starfish is to prototype a reference pipeline for the analysis of image-based transcriptomics data that works for each image-based transcriptomic assay. This is a work in progress and will be developed in the open.
We are currently in pre-alpha, finishing proof of concept pipelines for each of the spaceTx contributors that leverage starfish's shared object model. At this time starfish is mature enough to support computational developers interested in adapting other assays to starfish's object model.
| Assay | Loads Data | Single-FoV Pipeline | Multi-FoV Pipeline |
| MERFISH | [x] | [x] | in process |
| ISS | [x] | [x] | in process |
| osmFISH | [x] | in process | [ ] |
| allen_smFISH | [x] | in review | [ ] |
| BaristaSeq | [x] | in process | [ ] |
| DARTFISH | [x] | in review | [ ] |
| ex-FISH | [x] | [ ] | [ ] |
| StarMAP | [ ] | [ ] | [ ] |
| FISSEQ | no data | no pipeline | [ ] |
| seq-FISH | [ ] | [ ] | [ ] |
| Imaging Mass. Cyto. | [x] | [ ] | [ ] |
Concept
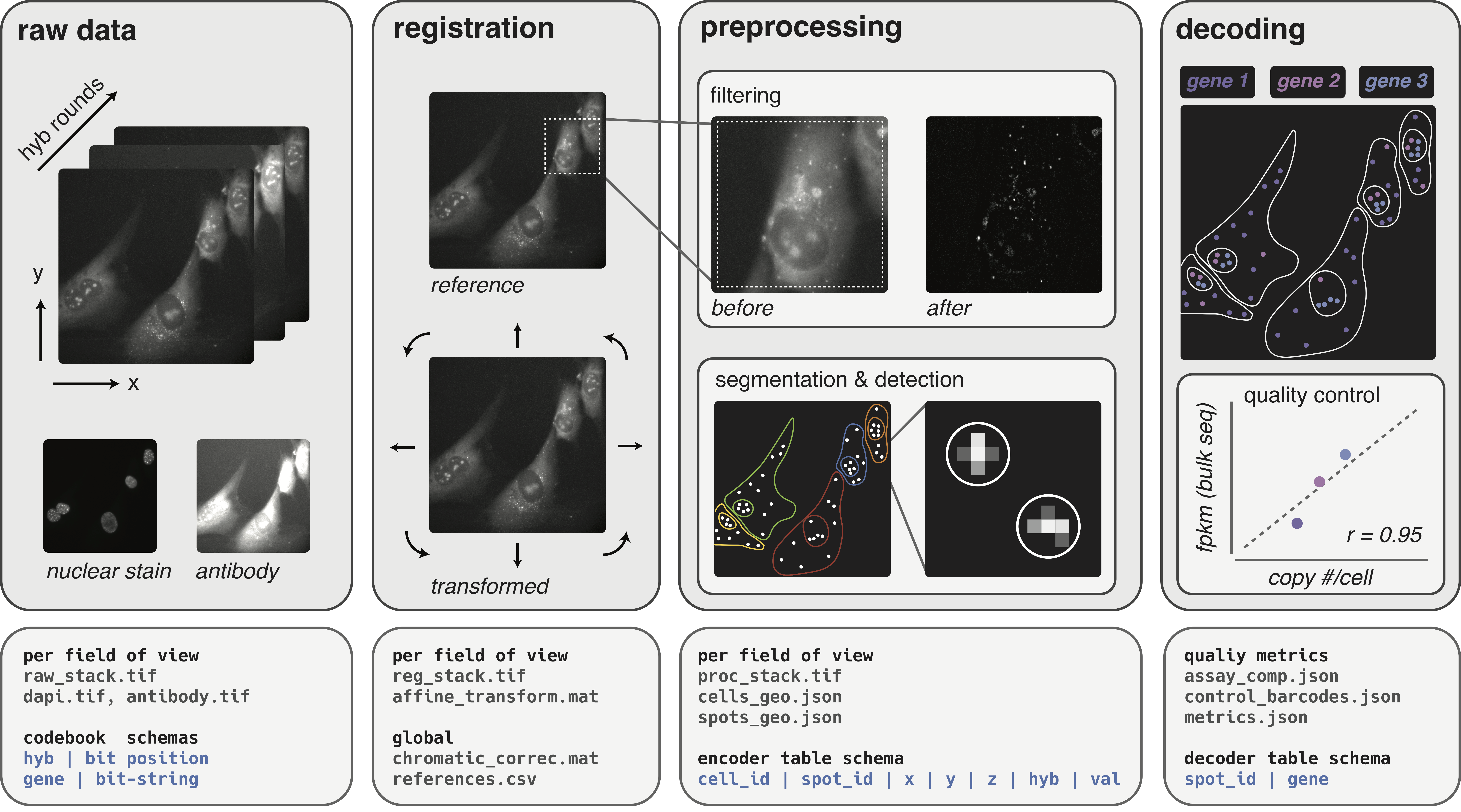
See this document for details. The diagram below describes the core pipeline components and associated file manifests that this package plans to standardize and implement.
Documentation
For more information on installation, usage, and the starfish API and CLI, please see the documentation