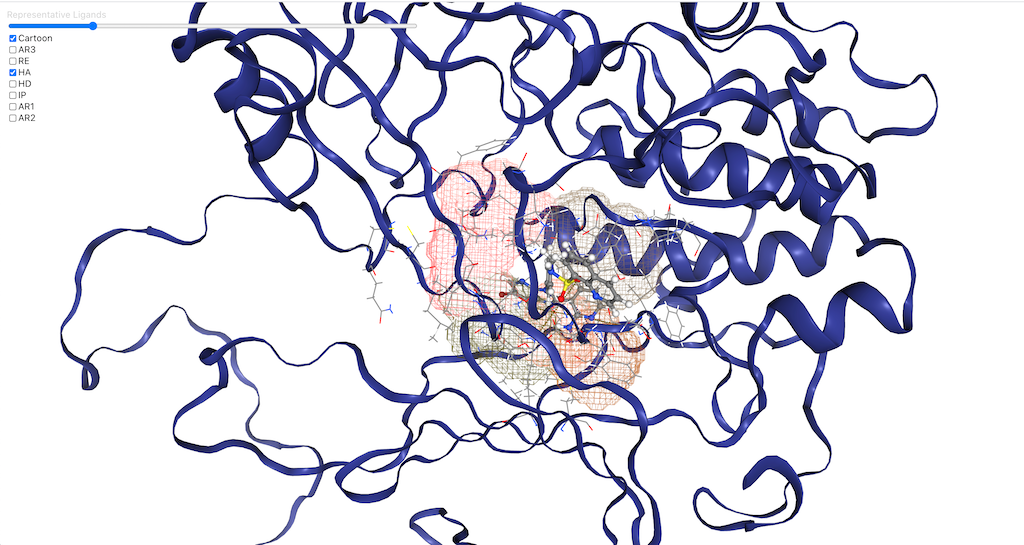
An visualization web app for the interactive design of drugs on Hypothesis & Branching platform.
hnbeye is an online React-based web app which will parse the input files/args from query parameters of request URI and dynamically render a page for further user interaction or embedded into other web services.
It has these main features:
- Pharmacophore Model Visualization: [?type=pharmod]
- Sankey Plot: [?type=sankey]
- Candidates Table View: [?type=candidates]
- Scaffold Graph View: [?type=scaffgraph]
- Protein Structure NGLView: [?type=protein]
The yarn and npm are required, here is the list of the installation manual:
- node >=v15.5.0 (includes npm >=7.4.0): https://nodejs.org/en/download/current/
- yarn >=1.22.10: https://classic.yarnpkg.com/en/docs/install/#mac-stable
After installation of npm and yarn, you can launch the demo through commands below:
yarn
yarn run start
All the visualization Component are listed under the /components/ folder. A typical way of extending a new component would have 2 steps:
import { React, Component } from 'react';
import { Stage } from 'ngl';
class CustomizedView extends Component {
static VIEW_TYPE = 'customized'
constructor(props) {
super(props);
this.state = {
queryParams: null,
argsURI: null,
args: null,
};
}
// Other source code ...
}
where 3 elements should be defined:
VIEW_TYPE: it will be used in App.js for parsing and initializing Component.queryParams: it has all the query parameters in the request URI as a dictionary.- Coding for view: your source code define the React Component acts as you expected.
The main branching of choosing which component to be rendered looks like the lines below:
if ('type' in query_params) {
const view_type = query_params['type']
if (view_type) {
if (view_type.toLowerCase() === PharModView.VIEW_TYPE) {
// e.g. ?pdb=/test/pharmod/5kcv-phar.pdb&sdf=/test/pharmod/5kcv-phar.sdf&type=pharmod
return <PharModView queryParams={query_params} />;
} else if (view_type.toLowerCase() === ScaffgraphView.VIEW_TYPE) {
return <ScaffgraphView queryParams={query_params} />;
} else if (view_type.toLowerCase() === ProteinView.VIEW_TYPE) {
return <ProteinView queryParams={query_params} />;
} else if (view_type.toLowerCase() === SankeyView.VIEW_TYPE) {
return <SankeyView queryParams={query_params} />;
} else if (view_type.toLowerCase() === CandidatesView.VIEW_TYPE) {
return <CandidatesView queryParams={query_params} />;
}
}
}
Define the extended VIEW_TYPE for your own component.