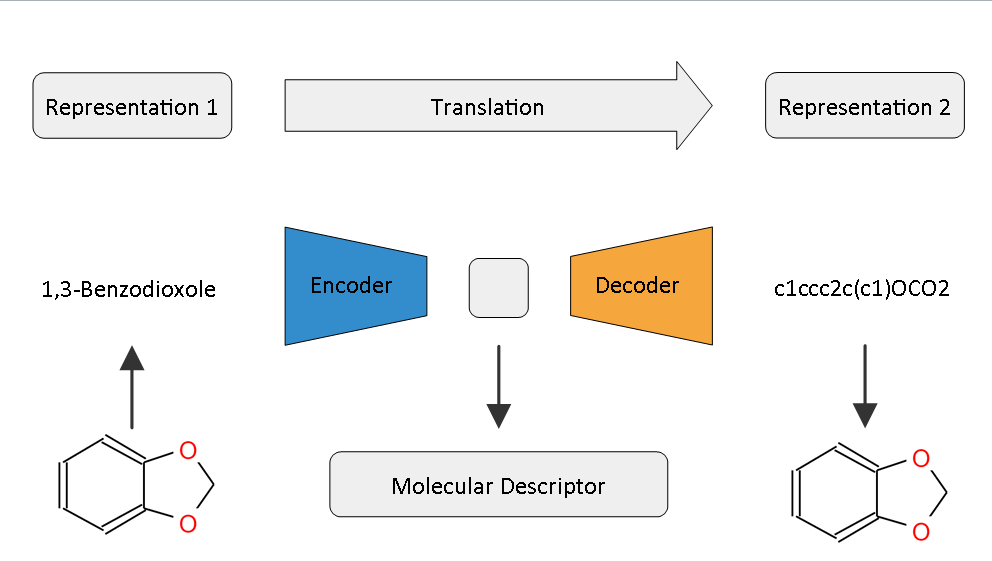
Implementation of the Paper "Learning Continuous and Data-Driven Molecular Descriptors by Translating Equivalent Chemical Representations" by Robin Winter, Floriane Montanari, Frank Noe and Djork-Arne Clevert.1
python 3
tensorflow 1.10
numpy
rdkit
scikit-learn
Create a new enviorment:
git clone https://github.com/jrwnter/cddd.git
cd cddd
conda env create -f environment.yml
source activate cdddInstall tensorflow without GPU support:
pip install --ignore-installed --upgrade https://storage.googleapis.com/tensorflow/linux/cpu/tensorflow-1.10.0-cp36-cp36m-linux_x86_64.whlOr with GPU support:
pip install --ignore-installed --upgrade https://storage.googleapis.com/tensorflow/linux/gpu/tensorflow_gpu-1.10.0-cp36-cp36m-linux_x86_64.whlAnd install the cddd package:
pip install .A pretrained model as described in ref. 1 is available on Google Drive. Download and unzip by execuiting the bash script "download_default_model.sh":
./download_default_model.shThe default_model.zip file can also be downloaded manualy under https://drive.google.com/open?id=1oyknOulq_j0w9kzOKKIHdTLo5HphT99h
Extract molecular descriptors from two QSAR datasets (ref. 2,3) and evaluate the perfromance of a SVM trained on these descriptors.
cd example
python3 run_qsar_test.py --model_dir ../default_modelor with gpu support on e.g. device 0:
python3 run_qsar_test.py --model_dir ../default_model --use_gpu --device 0
The accuracy on the Ames dataset should be arround 0.814 +/- 0.006.
The r2 on the Lipophilicity dataset should be arround 0.731 +/- 0.029.
Run the script run_cddd.py to extract molecular descripotrs of your provided SMILES:
cddd --input smiles.smi --output descriptors.csv --smiles_header smilesSupported input:
- .csv-file with one SMILES per row
- .smi-file with one SMILES per row
For .csv: Specify the header of the SMILES column with the flag --smiles_header (default: smiles)
The pretrained model can also be imported and used directly in python via the inference class:
import pandas as pd
from cddd.inference import InferenceModel
from cddd.preprocessing import preprocess_smilesLoad and preprocess data:
ames_df = pd.read_csv("example/ames.csv", index_col=0)
ames_df["smiles_preprocessed"] = ames_df.smiles.map(preprocess_smiles)
ames_df = ames_df.dropna()
smiles_list = ames_df["smiles_preprocessed"].tolist()Create a instance of the inference class:
inference_model = InferenceModel()Encode all SMILES into the continuous embedding (molecular descriptor):
smiles_embedding = inference_model.seq_to_emb(smiles_list)The infernce model instance can also be used to decode a molecule embedding back to a interpretable SMILES string:
decoded_smiles_list = inference_model.emb_to_seq(smiles_embedding)[1] R. Winter, F. Montanari, F. Noe and D. Clevert, Chem. Sci, 2019, https://pubs.rsc.org/en/content/articlelanding/2019/sc/c8sc04175j#!divAbstract
[2] K. Hansen, S. Mika, T. Schroeter, A. Sutter, A. Ter Laak, T. Steger-Hartmann, N. Heinrich and K.-R. MuÌ´Lller, J. Chem. Inf. Model., 2009, 49, 2077–2081.
[3] Z. Wu, B. Ramsundar, E. N. Feinberg, J. Gomes, C. Geniesse, A. S. Pappu, K. Leswing and V. Pande, Chemical Science, 2018, 9, 513–530.