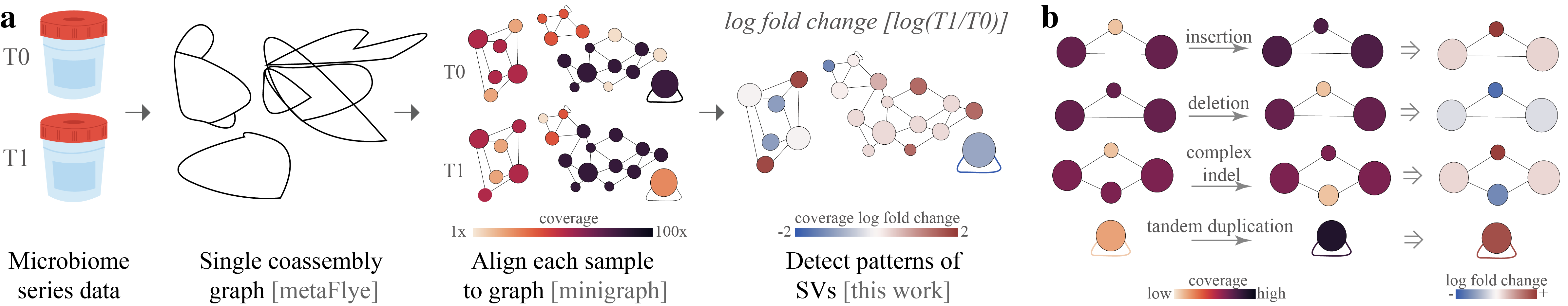
Rhea is a software used to detect structural variants (SVs) between steps in long-read metagenomic series data. Current SVs detected include: insertions, deletions, complex indel (insertion and deletion in the same location), and tandem duplications. Below is a graphic represtation of the rhea pipeline.
(a) To utilize rhea, first, microbiome series data must be collected and long whole genome sequencing reads generated. Then, within rhea, a coassembly graph of all reads in the series is created with metaFlye. Reads from each sample are then separately aligned to the coassembly graph with minigraph. Rhea evaluates log fold change in coverage between series steps for SV-specific patterns in the assembly graph to detect structural variants between steps. (b) Assembly graph patterns detected in rhea, which indicate potential insertions, deletions, complex indels, and tandem duplicates. Insertions and deletions are detected by observing a triangle where one node has a significantly higher (insertion) or lower (deletion) log fold change. Complex indels are noted by a square with one or two outliers; in the case of two outliers, the two outliers must be of opposing sides of the median and not have an edge between them. Tandem duplicates are detected by a log fold change of a self-loop edge coverage greater than 1.Here is a toy example to detect SV in two different variants E. coli in a metagenome.
First, download t0.fasta and t1.fasta from OSF under the example directory.
Then run rhea on the samples.
python ./rhea.py example/t0.fasta example/t1.fastaExpected output: All output files are expected to be in directory rhea_results.
Output files include: Bandage_metadata.csv, bp_counts.tsv, edge_coverage-t{0,1}, metaflye directory, node_coverage_norm.csv,
node_coverage.csv, rhea.log, structual_variants-c0, t{0,1}.gaf.
Expected run time: ~5 minutes for flye
Dependencies for rhea include: python v3.8+, numpy, pandas, networkx v3.2+, seaborn, Flye, and minigraph. Environment.yaml can be used build a conda environment with all necessary dependencies; rhea environment will need to be activated for each rhea run.
https://github.com/treangenlab/rhea.git
cd rhea
conda env create -f environment.yml
conda activate rheaWe highly recommend using metaFlye v2.9.3+ due to improvements in strain variations maintained.
structual_variants-c{1-N}.tsv: detected structual variants for each pair of subsequent samples. Denoted in the file name.node_coverage.csv: Spreadsheet where each row is a unique node. Contains sequence length for each node, detected coverage for each sample, linear change in coverage between subsequent pairs, and log fold change in coverage. Color hex values are also provided for each type of coverage/coverage difference for graph heatmap visuals.node_coverage_norm.csv: Same asnode_coverage.csv, except coverage values are normalized across all samples in the set based on total number of base pairs per sample. Use of normalized or raw values for SV detection is determined by the input parameter--raw-diff(normalized is default).edge_coverage-c{}.tsv: 2D matrix of edge coverage for each provided sample.Bandage_metadata.csv: consolidated data for regarding node length, coverage, coverage change, and detected SVs; designed to be used with Bandage to create visuals. See below.bp_counts.tsv: total number of bps in each sample; used for normalization.rhea.log: Computational log and established min/max values for heatmap graph visuals.
Rhea output can be used in conjunction with Bandage to see patterns SV and evolutionary activity in the metagenome.
- Install and open Bandage.
- Load assembly graph:
./rhea_results/metaflye/assembly_graph.gfa. - Select column for node colors from
Bandage_metadata.csvby changing any of the column name ending in_colourtocolour. Import CSVfor updated CSV into Bandage.- Draw graph
singlewithcustom colours. - Min/max color scale values can be found in rhea.log. Colored bar can be found in
docs/visual_scales.png.
| Command | Default | Description |
|---|---|---|
| input | (required) | paths to input metagenome sequences or alignments |
| --output-dir, -o | rhea_results | path to output directory |
| --type | nano-raw | type of reads for MetaFlye graph construction ['pacbio-raw', 'pacbio-corr', 'pacbio-hifi', 'nano-raw', 'nano-corr', 'nano-hq'] |
| --input-graph | (generated) | path to graph if alignments are provided (req. with alignments) |
| --bp-table | (generated) | path to bp counts per sample if alignments are provided (req. with alignments) |
| --node-std | 1 | number of standard deviations away from median to call indels |
| --edge-lfc-thresh | 1 | number of lfc increase to call duplications |
| --raw-diff | FALSE | set to true if no normalization for bp count between samples is desired |
| --collapse | FALSE | activate to collapse metaFlye bubble (i.e. not use --keep-haplotypes) |
| --flye-exec | flye | path to flye executable |
| --minigraph-exec | minigraph | path to minigraph executable |
| --threads, -t | 3 | number of threads utilized by flye & minigraph |
If you cite rhea, be sure to also cite metaFlye and mingraph. If you use rhea visuals, be sure to cite Bandage.
Rhea preprint: Curry, K. D., Yu, F. B., Vance, S. E., Segarra, S., Bhaya, D., Chikhi, R., Rocha, E. P. C., & Treangen, T. J. (2024). Reference-free Structural Variant Detection in Microbiomes via Long-read Coassembly Graphs (p. 2024.01.25.577285). bioRxiv. https://doi.org/10.1101/2024.01.25.577285
Validation tests & scripts: https://osf.io/fvhw8/