This python code is a rule-based NLP interpreter for automatically inferring sequence labels from variable natural text in DICOM files exported from Vancouver General Hospital. The features of this utility are:
- Automatically infer sequence types from natural text descriptions present in SeriesDescription tag in DICOM.
- Resample images to particular user defined spacing and target shape of the volume.
- Convert and save native DICOM files to any SimpleITK supported formats (*nii.gz, *mha, *mhd etc.)
The utility supports automatic detection of the following sequences:
- T1
- T2
- T1CE
- T2FLAIR
The assumptions and rules encoded in this utility are available in modules/helpers.py file.
Dependencies:
- [Required] Install python 3 on your system.
- Please refer to the instructions relevant to your system and user privileges.
- [Optional] Install virtualenvwrapper.
- Again, refer to official virtualenvwrapper instructions on their website.
Install required packages:
pip install -r requirements.txtor if you have sudo privileges:
sudo pip install -r requirements.txtPlace your exported DICOM files from VGH into a folder. The default Phillips PACS server folder structure for exported studies is:
<your_internal_path>/IMediaExport/DICOM
Use this as your --path argument.
Invoke the utility using:
python process_vgh_data.py \
--path="<path_to_dicom_files>" \
--save_path="default" \
--resample=1 \
--target_shape=240x240x155 \
--target_spacing=1x1x1 \
--verbose=0 \
--interpolator=0 \
--out_format="nii.gz"The command line arguments are:
--path: path to your exported DICOM files. Notice that the path points to the folder INSIDE which the patient folders (PAT_0000...) reside.--save_path: path to save converted/resampled files.--resample: whether or not to resample images. Supports resampling to a particular voxel spacing and shape.--target_shape: target shape if resampling is required, specified in format (WxHxZ).--target_spacing: target spacing if resampling is required, specified in format (SxSxS).--verbose: change verbosity level, 0 for info and 1 for debug. Warning: changing this to 1 will output a lot of logging information.--interpolator: which interpolator to use for resampling:- (0): BSpline
- (1): NearestNeighbor
- (2): Linear
--out_format: Output format to save files.
- Perform registration between sequences using a single sequence as template.
- Perform automatic skull stripping of sequences.
- Provide manual override interface for cases where automatic inference may fail to determine sequences.
- Add MM-GAN sequence synthesis feature to synthesize missing sequences.
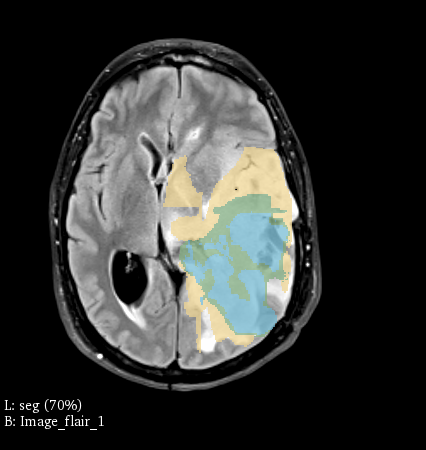
This utility may be used as a preliminary step for any pipeline that requires data to be in a structured and clean format. Typical use cases will be for machine learning/deep learning workflows which requires clean data in the form of sequences of particular shape and voxel spacing. In this project, a fully-working application is included in the form of a segmentation pipeline.
The segmentation pipeline is included in apps/segmentation-pipeline/ca-cnn and uses the open source pretrained models from https://github.com/taigw/brats17
Details regarding how segmentation is performed, and how the ca-cnn code is structured can be found on the original author's repository linked above.
The pipeline can be launched by the command:
bash run_pipeline.shHowever in order to run the pipeline, please ensure:
- You have a GPU enabled workstation.
- CUDA and CUDNN is installed and working.
- Created a new virtualenv using the
requirements.txtfile INSIDEca-cnnfolder.
The pipeline performs the following steps:
- Convert VGH DICOM data into nii.gz data with four sequence (T1, T2, T1CE, T2FLAIR)
- Copies the newly converted data into
apps/segmentation-pipeline/ca-cnn/data. - Creates a new folder
seg_outinapps/segmentation-pipeline/ca-cnn/data. - Prepares configuration file test_names.txt with the testing data present in
datafolder. - Loads pretrained BRATS2018 segmentation models from
model18into GPU memory. - Loads all data from
data. - Runs segmentation algorithm on loaded data.
- Write segmentation masks into
seg_outfolder for each patient.