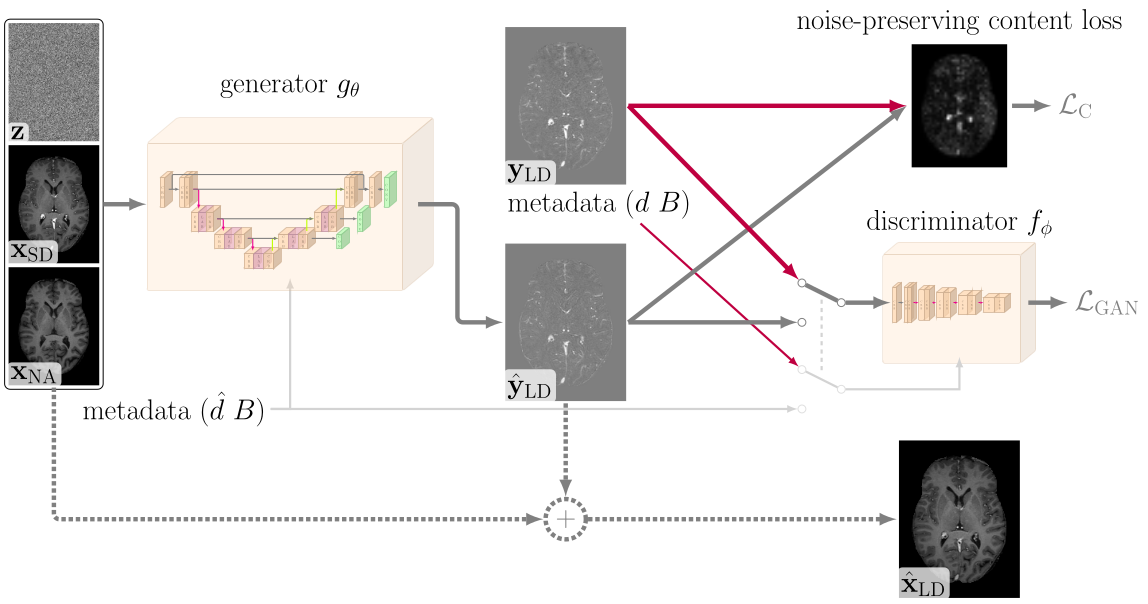
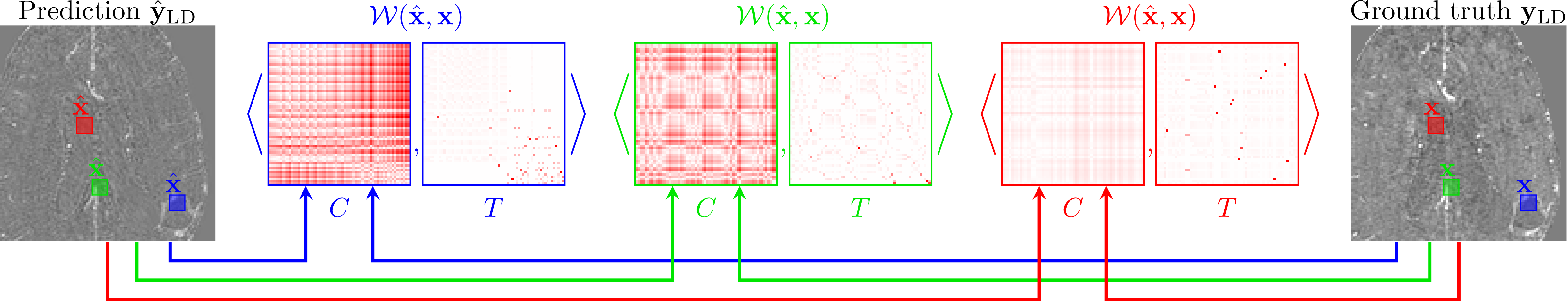
Faithful Synthesis of Low-dose Contrast-enhanced Brain MRI Scans using Noise-preserving Conditional GANs
Source code for the paper "Faithful Synthesis of Low-dose Contrast-enhanced Brain MRI Scans using Noise-preserving Conditional GANs".
Thomas Pinetz, Erich Kobler, Robert Haase, Katerina Deike-Hofmann, Alexander Radbruch, Alexander Effland
Institute of Applied Mathematics, University of Bonn, Germany
Department of Neuroradiology, University Medical Center Bonn, Germany
We provide a environment.yaml file to automatically build the environment.
To actually run the scripts the location of the execution should be inside the src folder.
For the generation of new low-dose images run for dose_level
python inference_brats.py --ds 0.2
To run the standard dose prediction run:
python brats_standard_dose_inference.py
For this script we expect the scans to already have been subject to pre-processing (e.g. spatial and radiometric registration), which is the case for the prediction produced by the inference_brats.py script.
We included the checkpoint used in the paper to generate the low-dose images and a checkpoint for
The proposed loss criterion is the function PatchWiseWasserSteinSinkhorn defined in loss_criterions.py.
We used the pytorch lightning framework to train our model and the base model is given in gan_models.
Network architectures are defined in networks_cond.py and discriminator.py and definition of hyperparameters are given in the respective config files.
In addition to our inhouse dataset, we also used the BraTS dataset to validate our method. We provide all necessary checkpoints and scripts to reproduce the results obtained on this publicly available dataset.
If you use the same dataset you should cite the respective paper as is descriped here: https://www.med.upenn.edu/cbica/brats2020/data.html
@article{pinetz2023faithful,
title={Faithful Synthesis of Low-dose Contrast-enhanced Brain MRI Scans using Noise-preserving Conditional GANs},
author={Pinetz, Thomas and Kobler, Erich and Haase, Robert and Deike-Hofmann, Katerina and Radbruch, Alexander and Effland, Alexander},
year={2023},
journal={arXiv preprint arXiv:2306.14678}
}