Predicting vegetation health from precipitation and temperature
This repository experiments with different machine learning models to predict drought indices in East Africa (specifically the Normalized Difference Vegetation Index) using temperature and precipitation data.
Models are trained on data before 2016, and evaluated on 2016 data. Vegetation health in June is being predicted.
In addition, vegetation health can be hidden from the model to better understand the effects of the other features.
| Model | RMSE | RMSE (no veg) |
|---|---|---|
| Linear Regression | 0.040 | 0.084 |
| Feedforward neural network | 0.038 | 0.070 |
| Recurrent neural network | 0.035 | 0.060 |
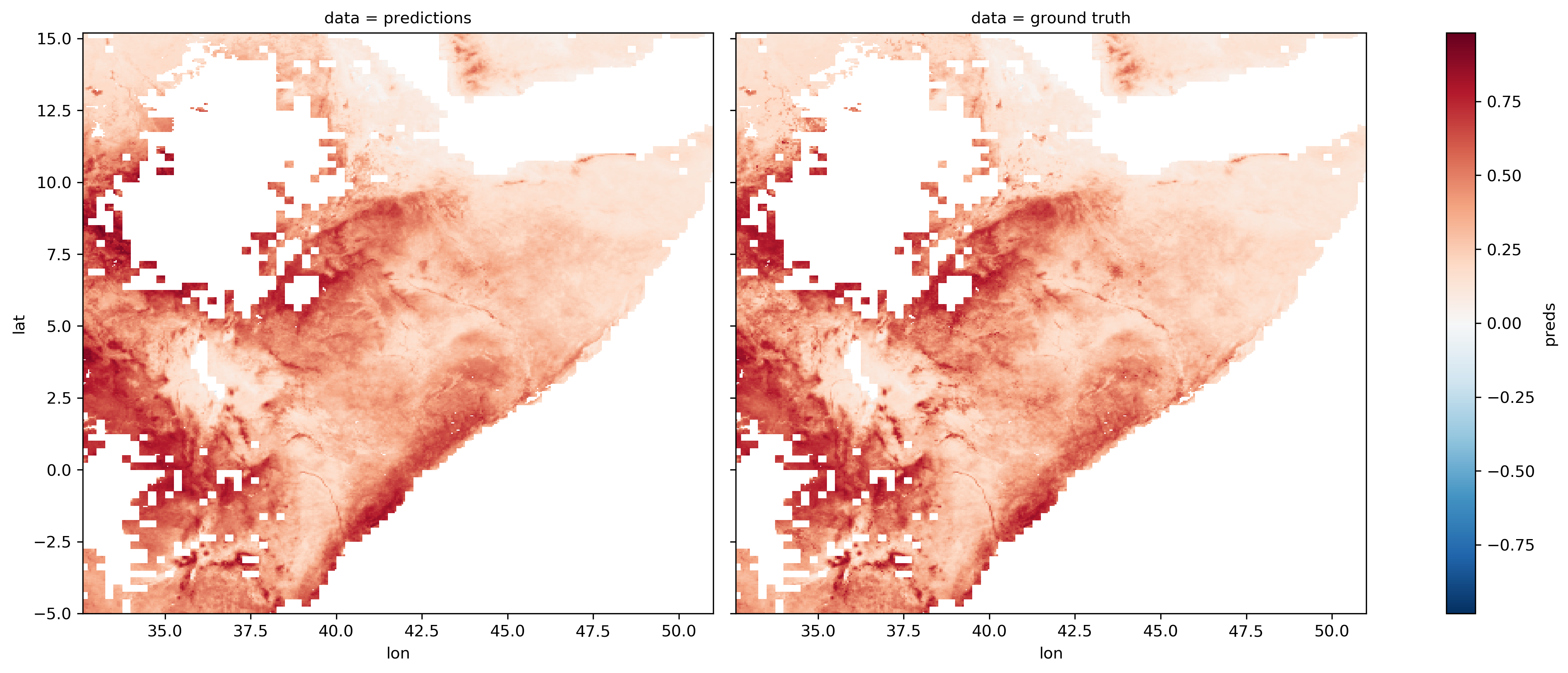
The results of the models can also be compared visually with the ground truths (the example below is from the baseline logistic regression):
In addition, the effects of the inputs on the models' predictions are investigated using shap values in Jupyter Notebooks, for both the feedforward neural network and the recurrent neural network.
Python Fire is used to generate a CLI.
Normalize values from the original csv file, remove null values, add a year series.
python run.py cleanA target can be selected by adding the flag --target, e.g. --target=ndvi_anomaly.
By default, the target is ndvi. The selected target must be in
predictor.preprocessing.VALUE_COLS.
The original data is currently generated using datasets on the Oxford University cluster, using the scripts
in data.
Turn the CSV into numpy arrays which can be input into the model.
python run.py engineer3 models have been implemented: a baseline linear regression, a feedforward neural network and a
recurrent neural network. They can be selected using the --model_type flag.
python run.py train_modelAnaconda running python 3.7 is used as the package manager. To get set up with an environment, install Anaconda from the link above, and (from this directory) run
conda env create -f environment.ymlThis will create an environment named vegetation_health with all the necessary packages to run the code. To
activate this environment, run
conda activate vegetation_health-
The following variables are used by the model:
['lst_night', 'lst_day', 'precip', 'sm', 'ndvi', 'evi', 'ndvi_anomaly']. They are all from different sources -
East Africa is defined here as the area of the original
.ncfile (spi_spei.nc)lat min, lat max :
-4.9750023,15.174995lon min, lon max :
32.524994,48.274994This makes the following bounding box: (left, bottom, right, top):
(32.524994, -4.9750023, 15.174995, 48.274994)