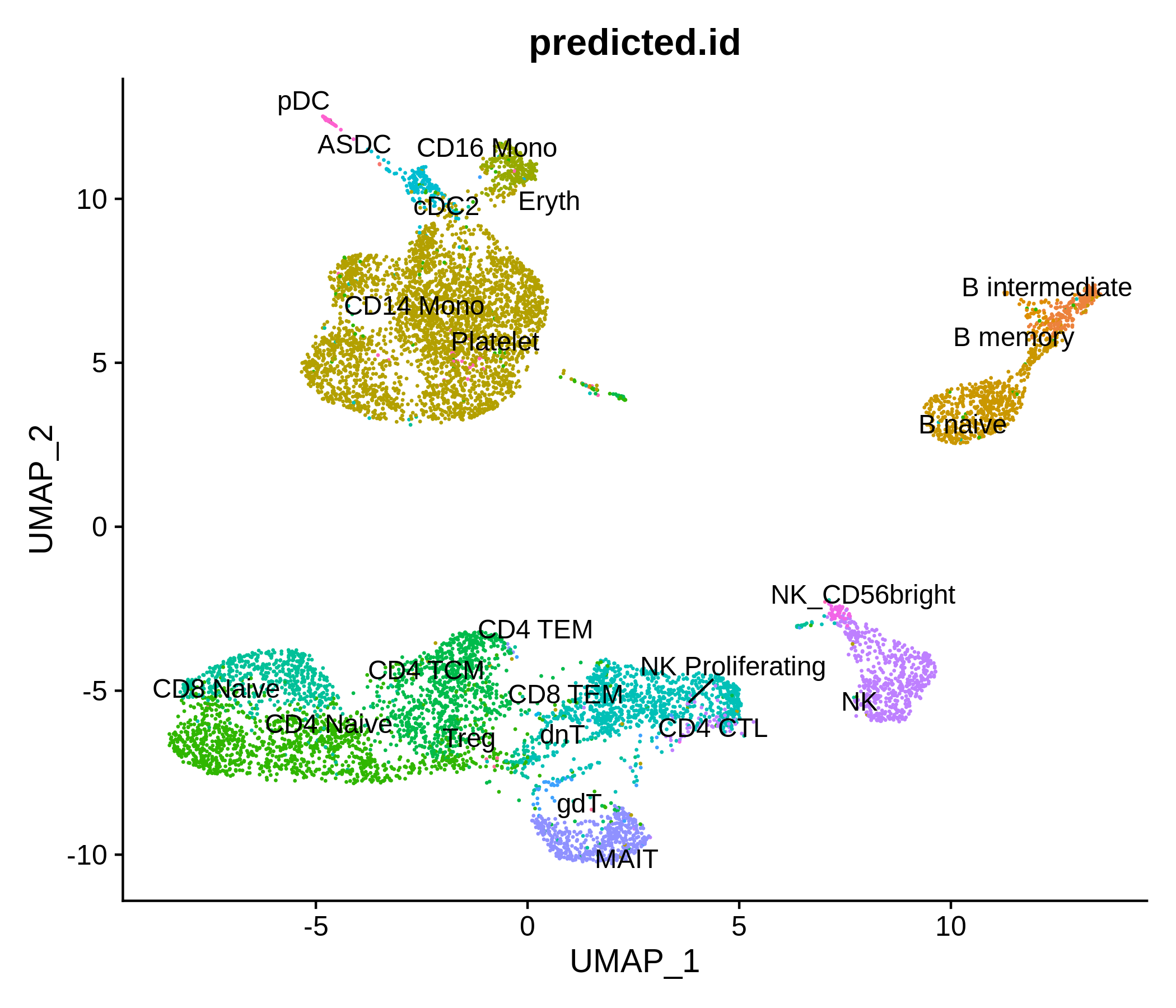
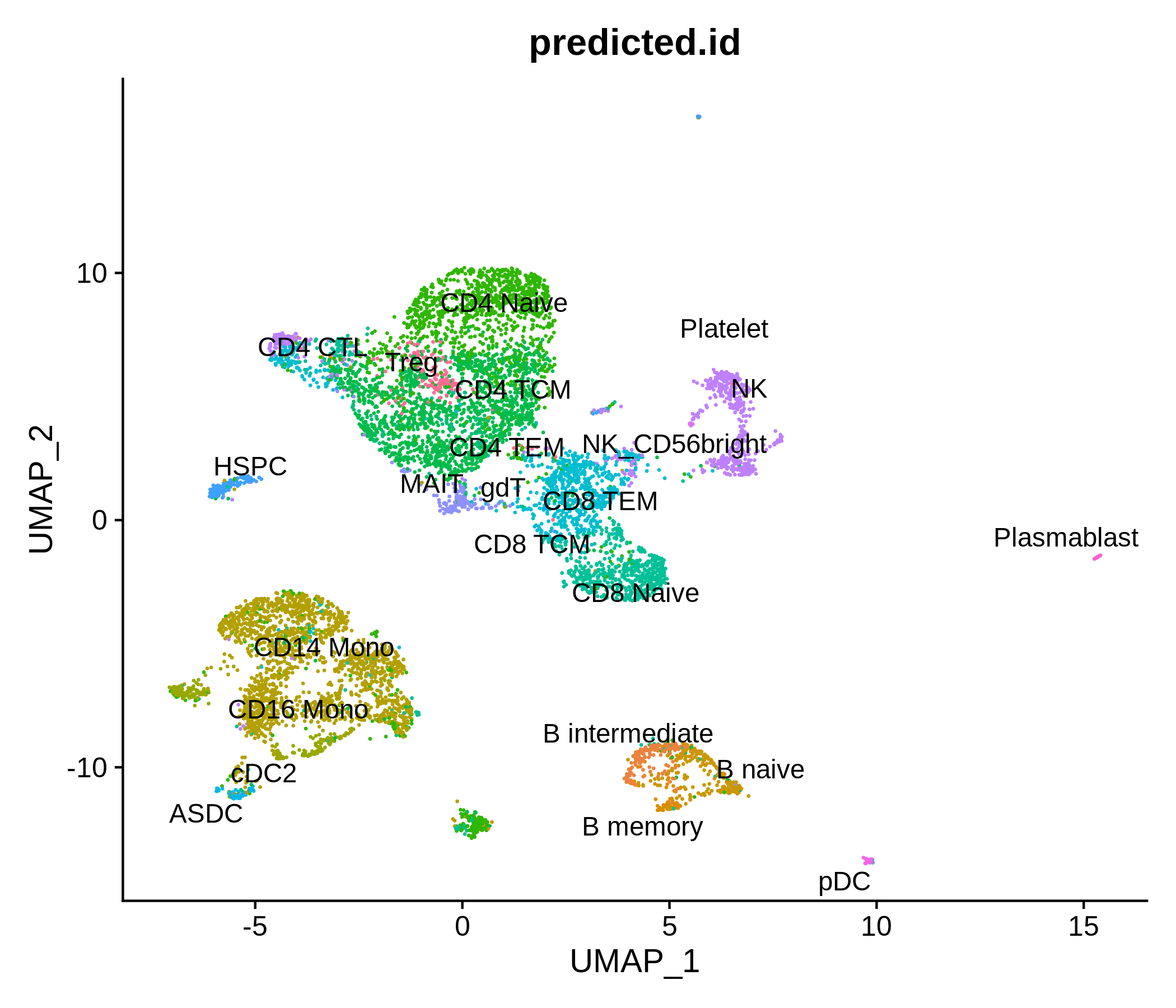
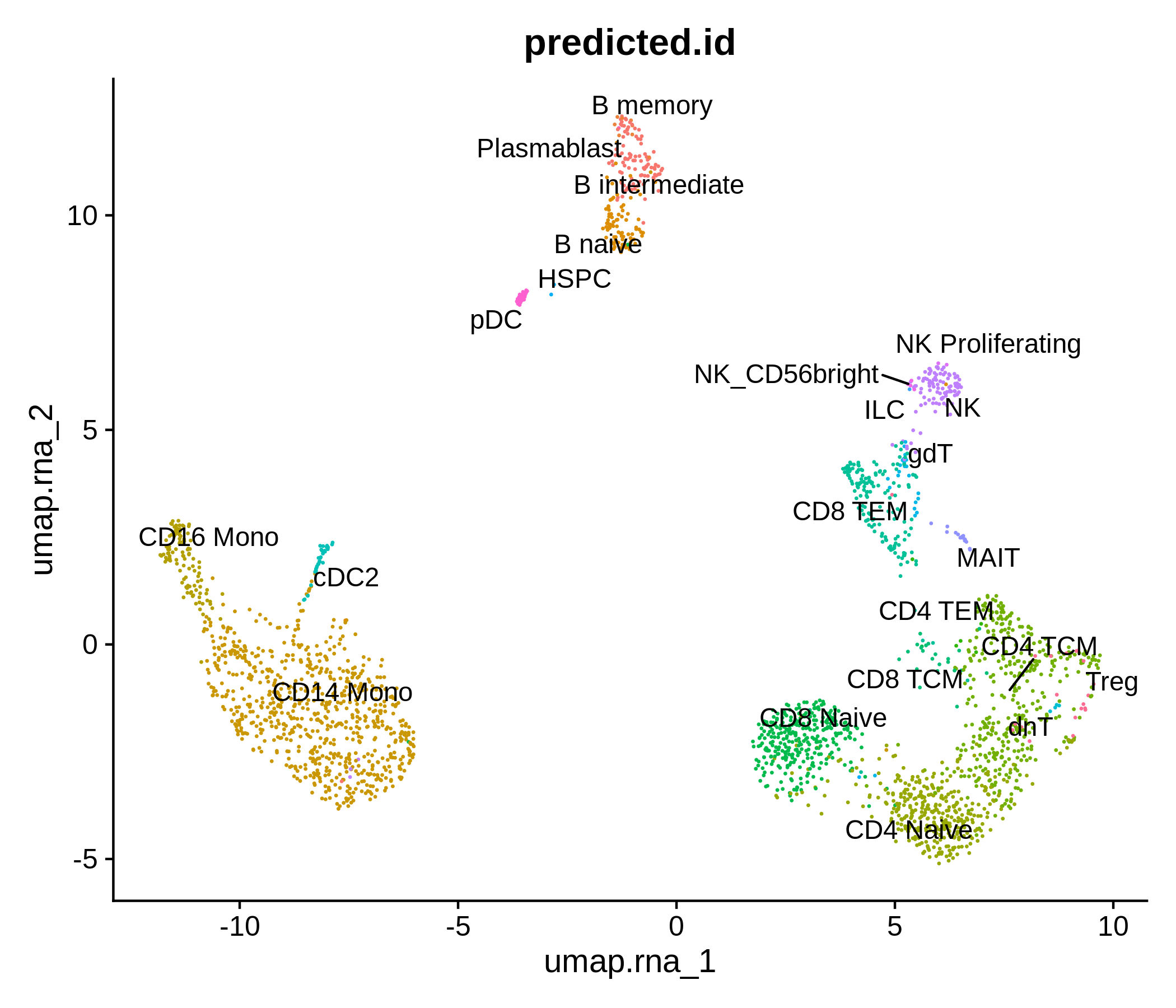
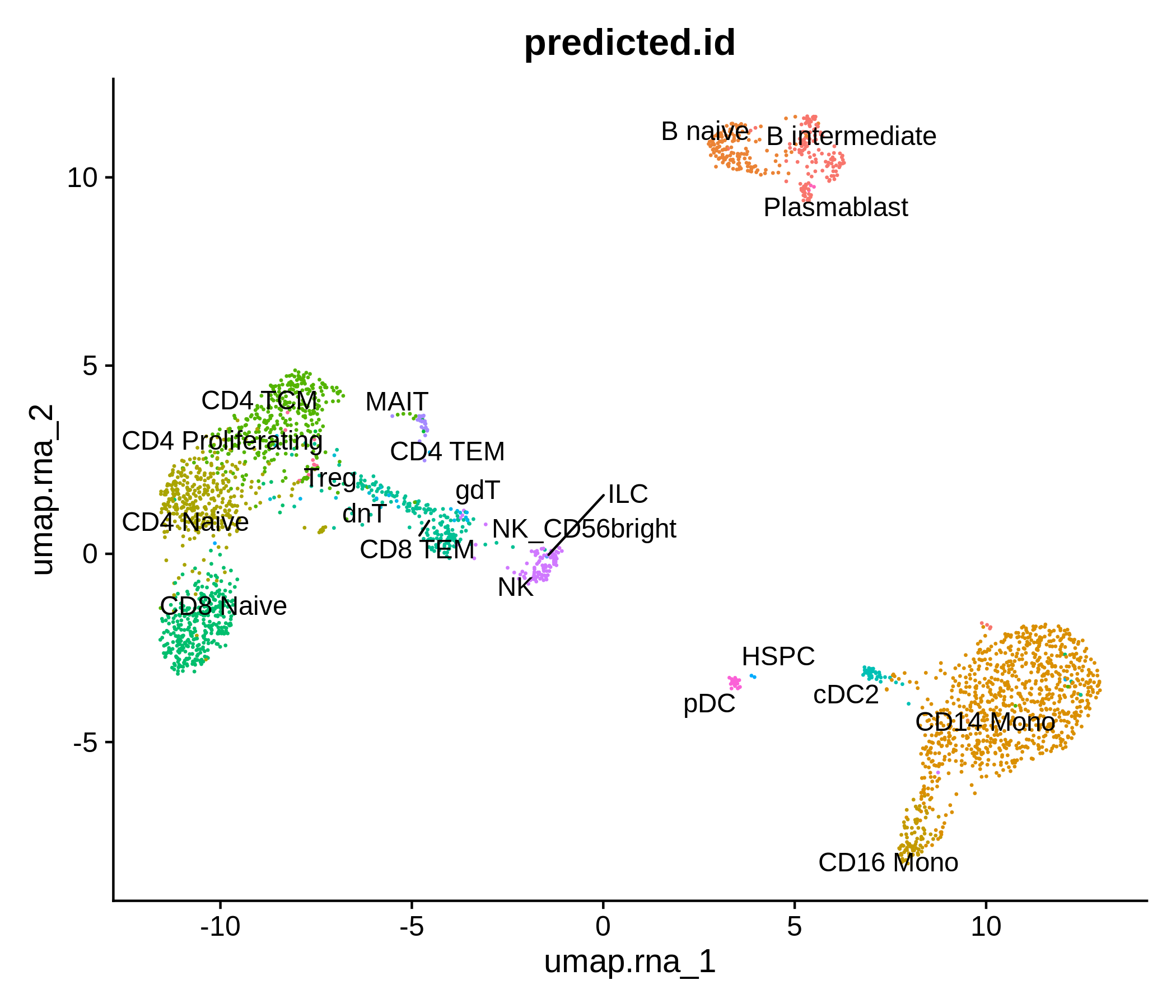
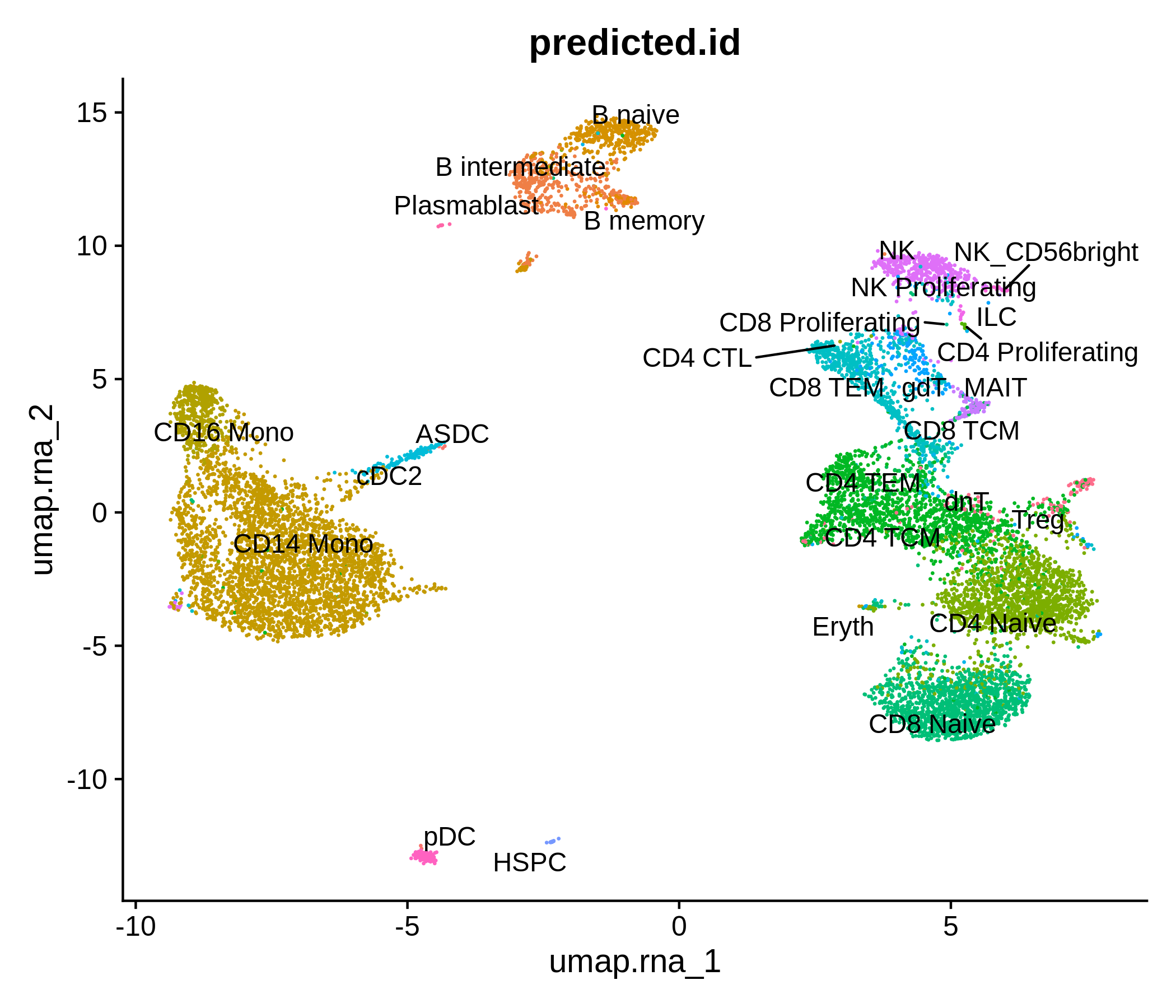
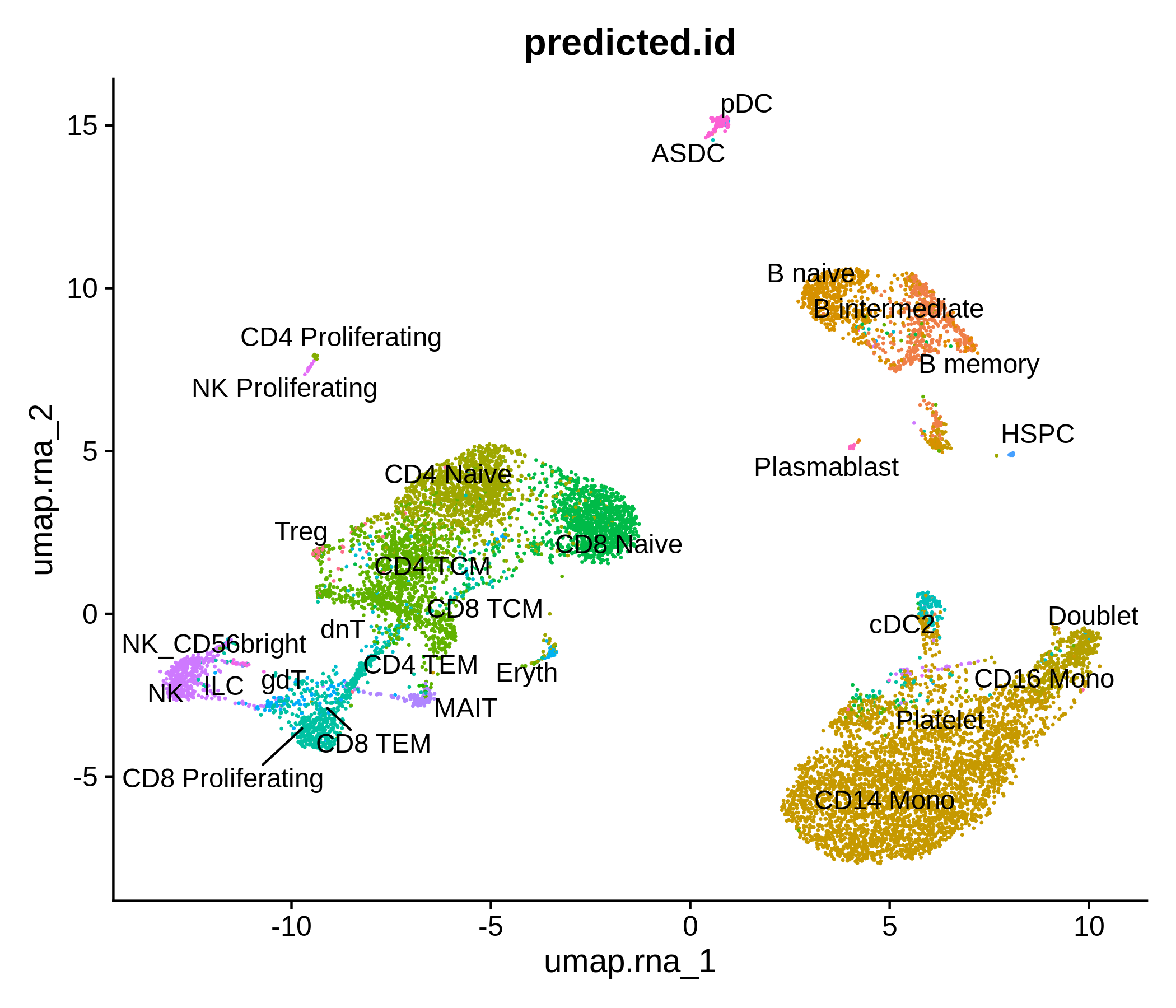
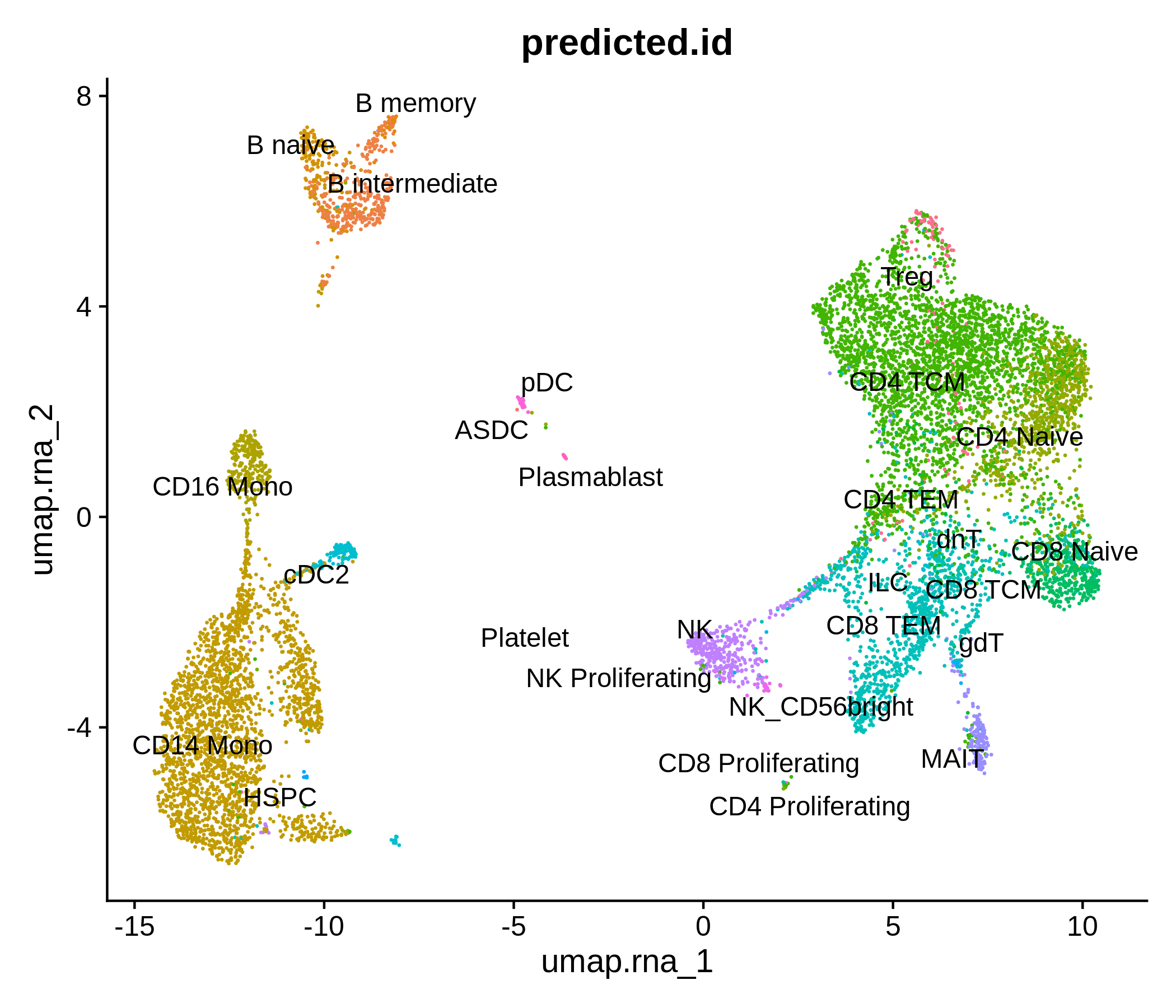
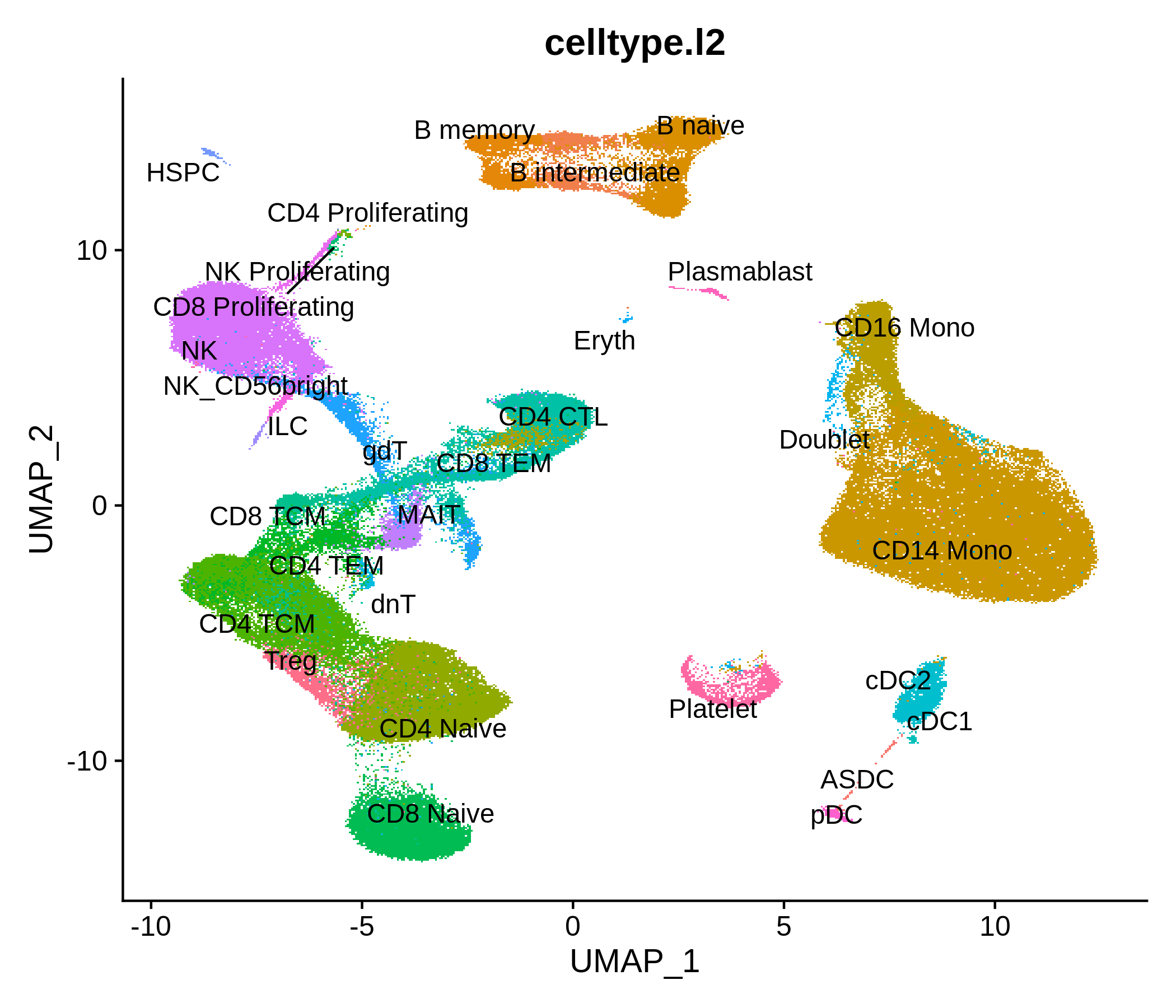
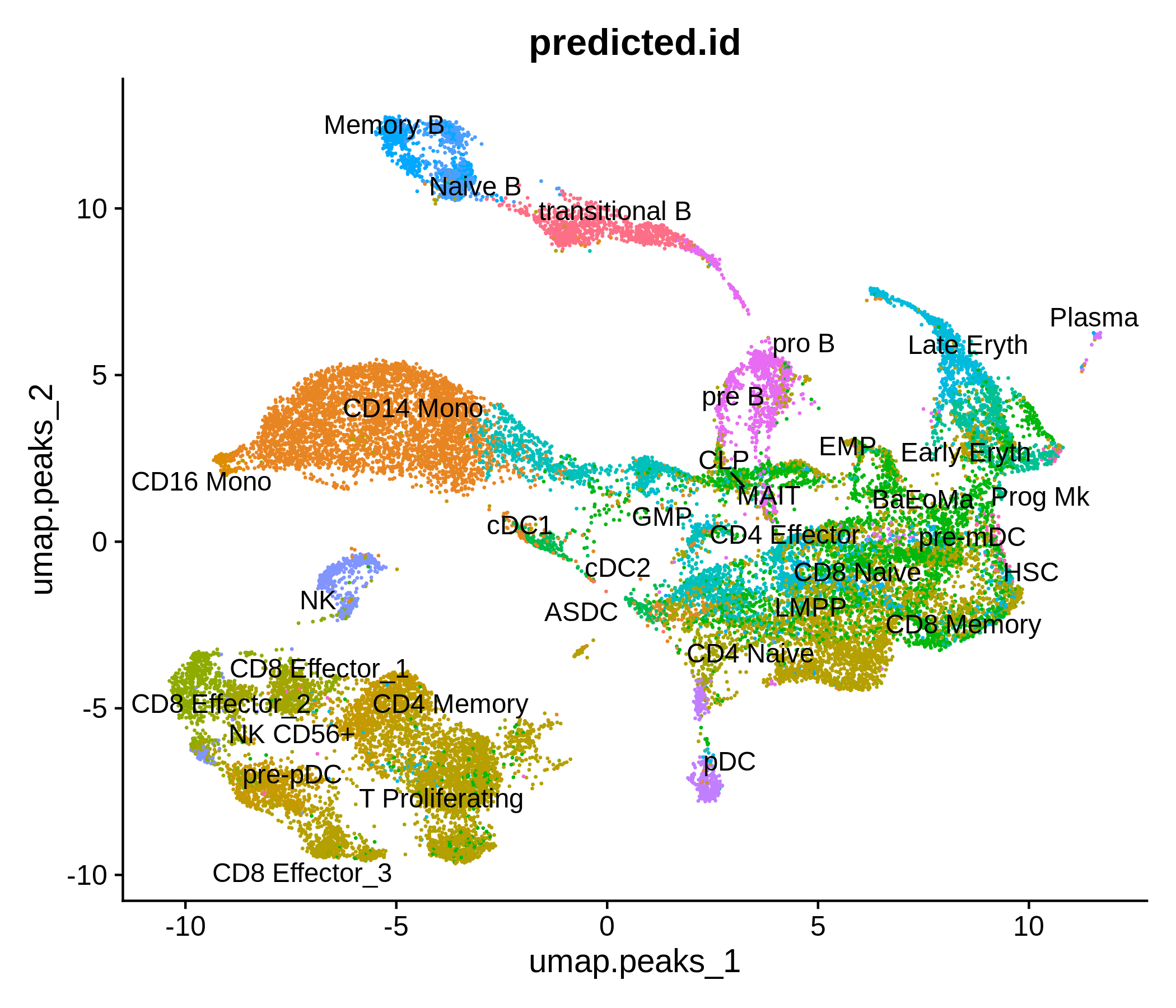
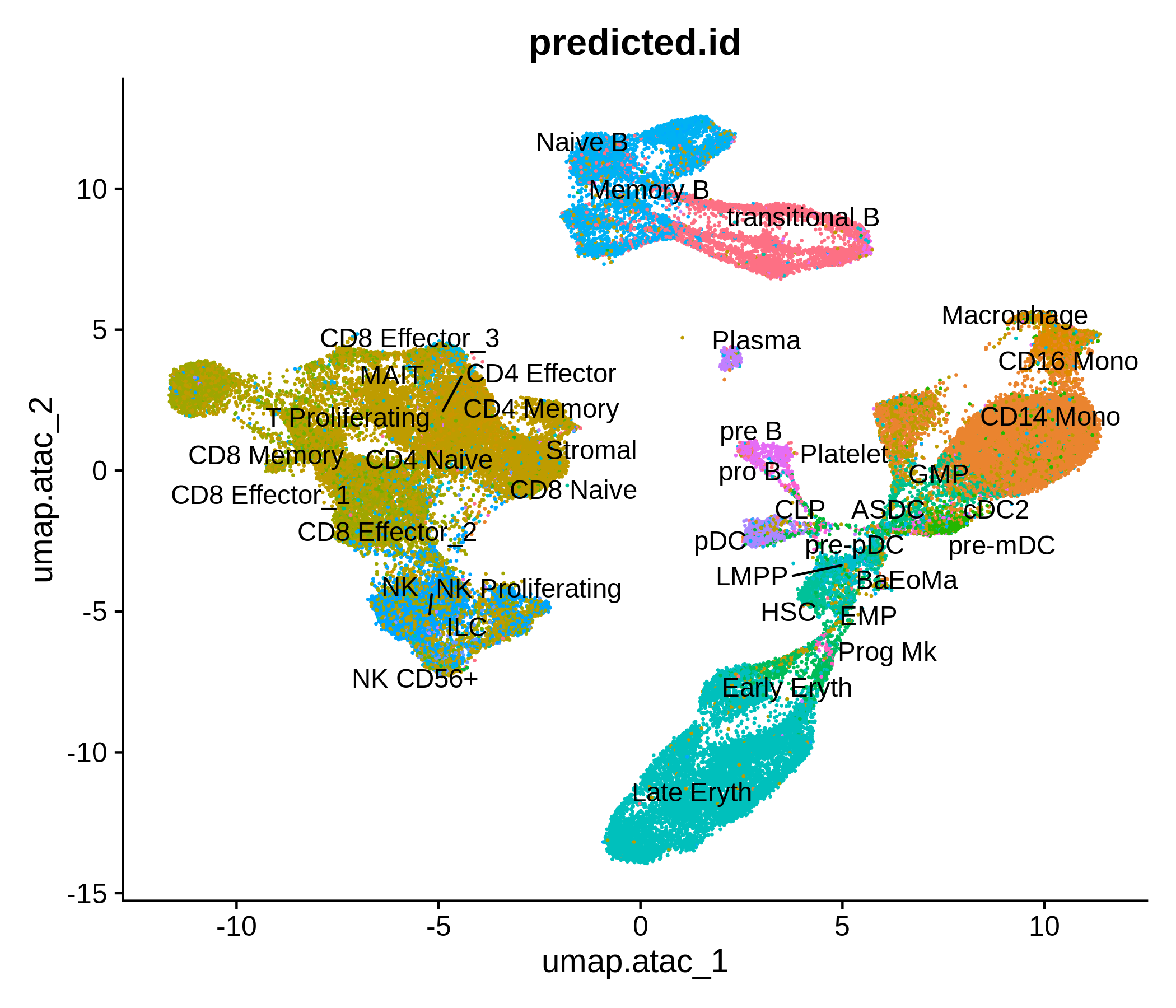
Processing pipeline for public single-cell epigenomic immune datasets.
All the required dependencies needed to run the workflow can be installed automatically by creating a new conda environment.
First ensure that conda or mamba is installed and available.
To create a new environment with the dependencies installed, run:
# using conda
conda env create -f environment.yaml
# using mamba
mamba env create -f environment.yaml
This workflow involves downloading data from AWS using the AWS
command line tools. To enable the download, you will first need
to create an AWS account and set up the AWS command line tools by
running aws configure. Note that some of the data downloaded
may incur charges from AWS.
To run the Snakemake workflow, first activate the conda environment containing the required dependencies:
conda activate immune
Next, run snakemake with the desired options. Setting the -j parameter
controls the maximum number of cores used by the workflow:
snakemake -j 24
See the snakemake documentation for a complete list of available options.