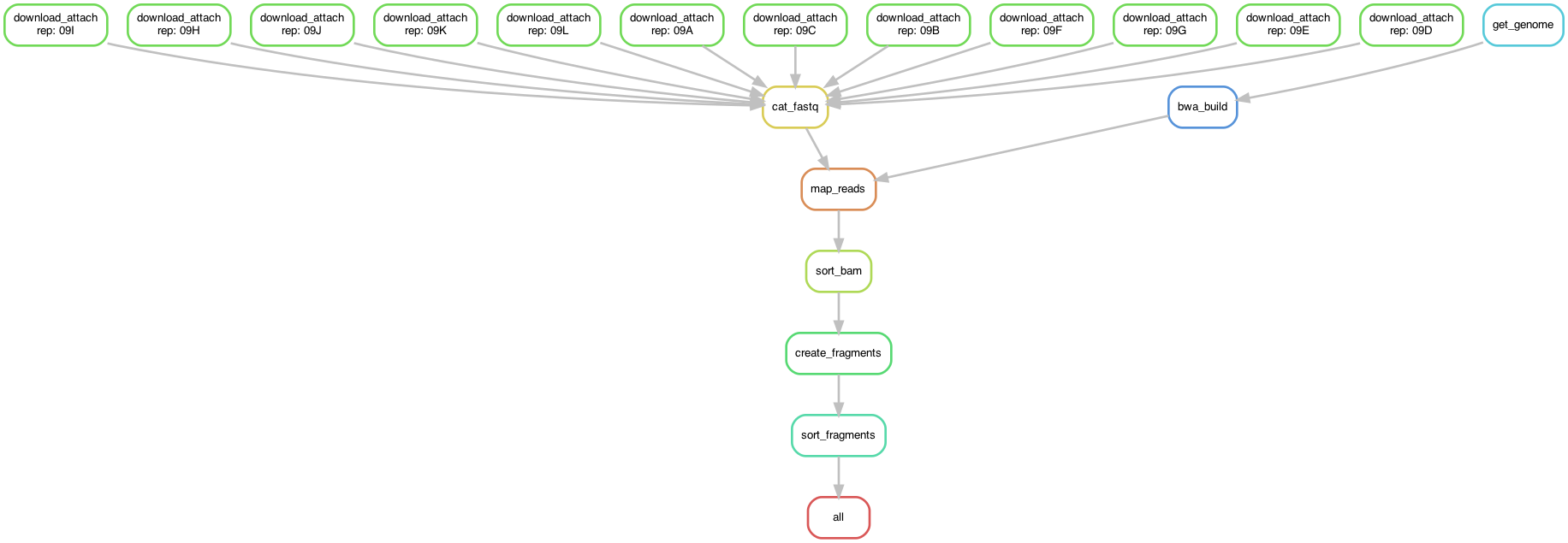
Code to process raw SNARE-seq ATAC reads for the adult mouse brain.
Steps:
- Download fastq files from SRA for each replicate (12 replicates)
- Attach cell barcode and replicate prefix to fastq read name using sinto
- Download mm10 genome fasta from UCSC and create bwa index
- Map fastq files to mm10 using bwa mem, pipe output to samtools to create bam file
- Sort and index bam file with samtools
- Create fragment file from bam file using sinto
- Sort, bgzip-compress, and index fragment file using bgzip and tabix
The pipeline is constructed using snakemake. To run the pipeline, first install miniconda, then create a conda environment for the pipeline using the environment.yaml file. This will install all the dependencies needed:
conda env create -f environment.yaml
This creates an environment called snareseq. Activate this environment when running the
pipeline: conda activate snareseq.
You can execute a dry-run of the pipeline to see what steps will be run using:
snakemake -n
To run the pipeline and generate the outputs, run:
snakemake --cores 8
Note that this involves >500 Gb of raw data.
Overview of the pipeline: