Author: Ji Huang
Date: 2023-10-31
Last update: 2023-11-17
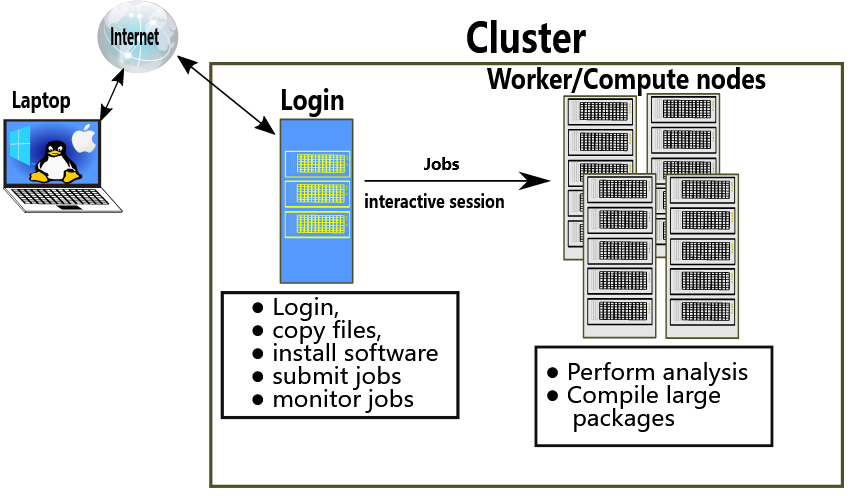
You will need a terminal to interact with the server. Many cases, you will also need a FTP client to upload/download files to the server.
On windows, I like: Windows Terminal, Putty, MobaXterm, WinSCP, and FileZilla.
tmux is very good helper.
You can set up the ssh key for a faster and better connection to the server.
You will need a text editor to create/modify files on the server.
I like: Vim, VScode and Sublime Text.
Helper: find .vscode-server/ -type f | wc -l
Once you generate the expression count matrix, you will mostly work in R for the RNA-Seq analysis.
RStudio, RStudio server, Jupyter Notebook.
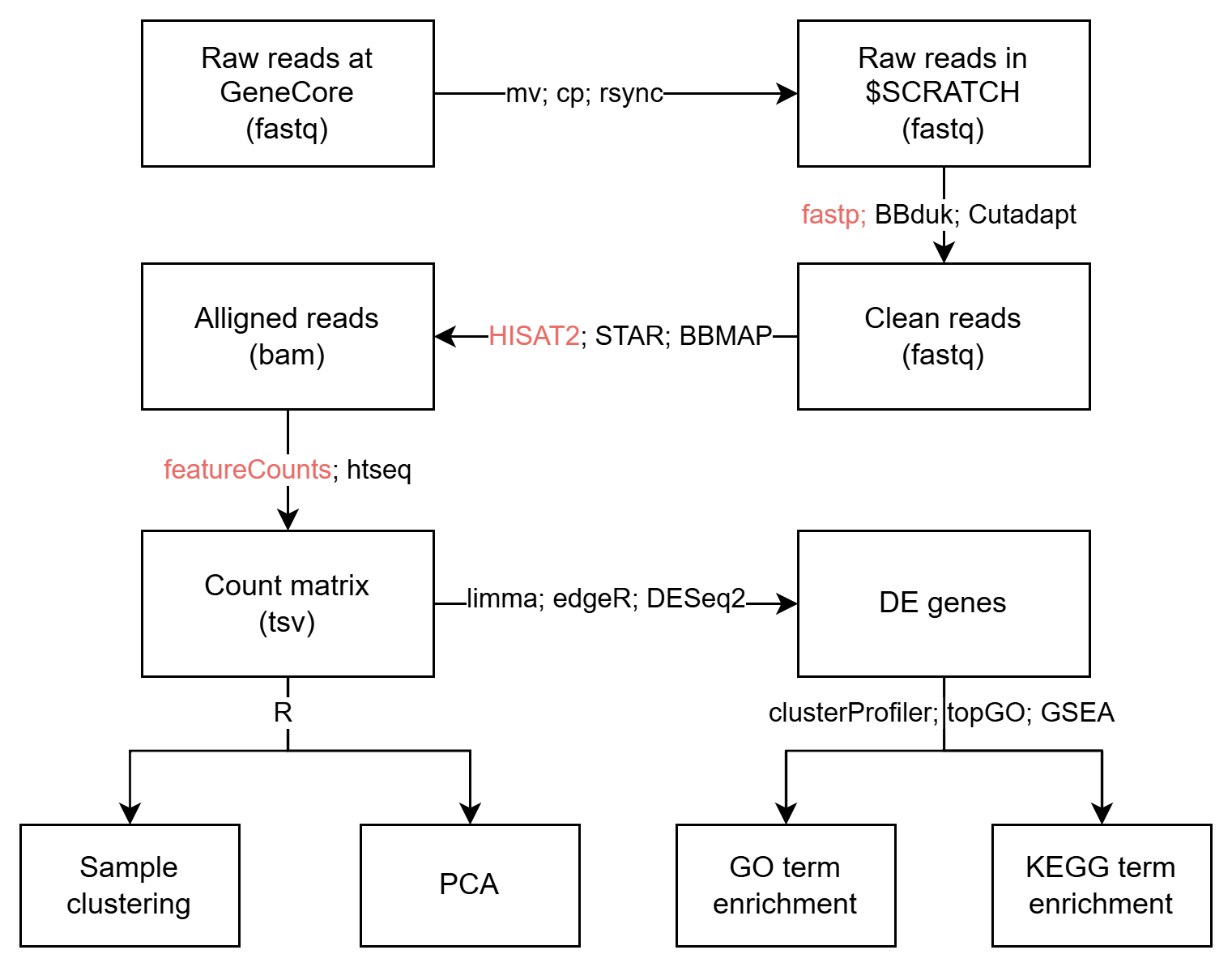
The Basic pipeline that works with most of the RNA-seq data without UMI.
Some slurm command that I use:
1. srun --mem 4GB --cpus-per-task 1 -t02:00:00 --pty /bin/bash
2. sacct -j <jobid> --format=JobID,JobName,state,exitcode,derivedexitcode,MaxRSS,Elapsed,MaxVMSize,MaxVMSizeNode,ReqMem
3. scontrol show jobid -dd <jobid> # for job details
4. seff <jobid> # efficiency of resource usage by the completed jobA simple slurm job header:
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=1 # modify this
#SBATCH --time=1:00:00 # modify this
#SBATCH --mem=2GB # modify this
#SBATCH --job-name=test # modify this
#SBATCH --mail-type=NONE
#SBATCH --mail-user=whoever@nyu.edu
#SBATCH --output=slurm_%j.out
module purge
All for single-end reads
# cleaning
fastp -l 20 --thread 1 -y -t 1 -x -a AGATCGGAAGAGC -f 2 \
-i input.fq.gz -o output2.fq.gz;
# aligning
hisat2 -p 2 -x ${REF} -U output2.fq.gz 2>aln.info | \
samtools view -bSh - | samtools sort -o output2.bam -
# counting
featureCounts -s 1 -T 2 -a $GTF -o final.feacureCounts output2.bamfastqformat. Wiki
A FASTQ file has four line-separated fields per sequence:
Field 1 begins with a '@' character and is followed by a sequence identifier and an optional description (like a FASTA title line).
Field 2 is the raw sequence letters.
Field 3 begins with a '+' character and is optionally followed by the same sequence identifier (and any description) again.
Field 4 encodes the quality values for the sequence in Field 2, and must contain the same number of symbols as letters in the sequence.
>@SRR8699958.1 1/1
CGGGACTATACATTTACAACAAAAAGAAACAAATCTTGTGGTCAAAGTTTCCATACGTAGCTTCTCTTCTCTACAC
+
AAA/A/EAE6AAAE/EEEA/EEEEE6AEEEEAE6/E/////EEEEE6E/E/EA/EA//E/6/6EEEEE/E<EAE/6
#!genome-build TAIR10
#!genome-version TAIR10
#!genome-date 2008-04
#!genome-build-accession GCA_000001735.1
#!genebuild-last-updated 2010-09
1 araport11 gene 10942648 10944727 . - . gene_id "AT1G30814"; gene_name "AT1G30814"; gene_source "araport11"; gene_biotype "protein_coding";
1 araport11 transcript 10942648 10944727 . - . gene_id "AT1G30814"; transcript_id "AT1G30814.1"; gene_name "AT1G30814"; gene_source "araport11"; gene_biotype "protein_coding"; transcript_name "AT1G30814-203"; transcript_source "araport11"; transcript_biotype "protein_coding"; tag "Ensembl_canonical";
1 araport11 exon 10944317 10944727 . - . gene_id "AT1G30814"; transcript_id "AT1G30814.1"; exon_number "1"; gene_name "AT1G30814"; gene_source "araport11"; gene_biotype "protein_coding"; transcript_name "AT1G30814-203"; transcript_source "araport11"; transcript_biotype "protein_coding"; exon_id "AT1G30814.1.exon1"; tag "Ensembl_canonical";
1 araport11 exon 10944078 10944229 . - . gene_id "AT1G30814"; transcript_id "AT1G30814.1"; exon_number "2"; gene_name "AT1G30814"; gene_source "araport11"; gene_biotype "protein_coding"; transcript_name "AT1G30814-203"; transcript_source "araport11"; transcript_biotype "protein_coding"; exon_id "AT1G30814.1.exon2"; tag "Ensembl_canonical";
1 araport11 CDS 10944078 10944225 . - 0 gene_id "AT1G30814"; transcript_id "AT1G30814.1"; exon_number "2"; gene_name "AT1G30814"; gene_source "araport11"; gene_biotype "protein_coding"; transcript_name "AT1G30814-203"; transcript_source "araport11"; transcript_biotype "protein_coding"; protein_id "AT1G30814.1"; tag "Ensembl_canonical";
@HD VN:1.0 SO:coordinate
@SQ SN:1 LN:30427671
@SQ SN:2 LN:19698289
@SQ SN:3 LN:23459830
@SQ SN:4 LN:18585056
@SQ SN:5 LN:26975502
@SQ SN:Mt LN:366924
@SQ SN:Pt LN:154478
@PG ID:hisat2 PN:hisat2 VN:2.2.1 CL:"/share/apps/hisat2/2.2.1/hisat2-align-s --wrapper basic-0 -p 2 -x /scratch/jh6577/tutorial_workshop/genome/genome_ath_trans --read-lengths 73,68,64,67,63,65,66,62,61,56,60,59,57,49,48 -U /tmp/2195840.unp"
@PG ID:samtools PN:samtools PP:hisat2 VN:1.12 CL:samtools view -bSh -
@PG ID:samtools.1 PN:samtools PP:samtools VN:1.12 CL:samtools sort -o DIV1_p1.bam -
@PG ID:samtools.2 PN:samtools PP:samtools.1 VN:1.12 CL:samtools view -h DIV1_p1.bam
SRR8699970.696 0 1 3904 60 10M82N63M * 0 0 AACTTGCGCTTCCAGTCAAAGTACAAATCGAGAGATGCTATGTGGTACTTCTTCTCTCGTAGAGAAAACAACA AAAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:0 MD:Z:73 YT:Z:UU XS:A:+ NH:i:1
SRR8699970.3584 0 1 4216 60 61M209N12M * 0 0 CAAAACCAAATCTGATTGGGTTATCCACGAGTTCCACTACGACCTCTTACCAGAACATCAGAGGACATATGTC AAAE6AAEEAEEE/EEEEAEE/EE/AEEAEAEE/EE</A<E<A<EEEEAEEEAEAEEEEE/AEEEEEEEEAE< AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:0 MD:Z:73 YT:Z:UU XS:A:+ NH:i:1
SRR8699970.5292 0 1 4855 60 73M * 0 0 TTTTGCAAATCACGGCGGTCAGTGGCTGAGTGACTATATCGACCTGCAACAGCAAGTTCCTTACTTGGCACCT AAAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE<EEE AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:0 MD:Z:73 YT:Z:UU NH:i:1
Download data, only use 10000 reads for demonstration, clean reads.
# https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117857
## Download fastq files
wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR869/000/SRR8699970/SRR8699970.fastq.gz
wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR869/008/SRR8699958/SRR8699958.fastq.gz
## Change name
mv SRR8699970.fastq.gz DIV1_rep1.fastq.gz
mv SRR8699958.fastq.gz EV_rep1.fastq.gz
## Only use 10000 reads
zcat DIV1_rep1.fastq.gz |head -n 40000 | gzip > DIV1_p1.fq.gz
zcat EV_rep1.fastq.gz |head -n 40000 | gzip > EV_p1.fq.gzClean reads
## Clean reads
module load fastp/intel/0.20.1
fastp -l 20 --thread 1 -y -t 1 -x -a AGATCGGAAGAGC -f 2 -i DIV1_p1.fq.gz -o DIV1_p1_clean.fq.gz;
fastp -l 20 --thread 1 -y -t 1 -x -a AGATCGGAAGAGC -f 2 -i EV_p1.fq.gz -o EV_p1_clean.fq.gz;Download Arabidopsis genome DNA sequence and GTF annotation.
## Continue with HISAT2-build
wget https://ftp.ebi.ac.uk/ensemblgenomes/pub/release-57/plants/fasta/arabidopsis_thaliana/dna/Arabidopsis_thaliana.TAIR10.dna.toplevel.fa.gz
wget https://ftp.ebi.ac.uk/ensemblgenomes/pub/release-57/plants/gtf/arabidopsis_thaliana/Arabidopsis_thaliana.TAIR10.57.gtf.gz
gzip -d Arabidopsis_thaliana.TAIR10.dna.toplevel.fa.gz;
mv Arabidopsis_thaliana.TAIR10.dna.toplevel.fa genome.fa
gzip -d Arabidopsis_thaliana.TAIR10.57.gtf.gz
mv Arabidopsis_thaliana.TAIR10.57.gtf genome.gtf
module load hisat2/2.2.1
hisat2_extract_splice_sites.py genome.gtf > genome.ss
hisat2_extract_exons.py genome.gtf > genome.exonPrepare a slurm job for building genome index for HISAT2.
hisat2_build.slurm
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=4 # modify this
#SBATCH --time=00:30:00 # modify this
#SBATCH --mem=8GB # modify this
#SBATCH --job-name=hisat2-build # modify this
#SBATCH --mail-type=NONE
#SBATCH --mail-user=whoever@nyu.edu
#SBATCH --output=slurm_%j.out
module purge
module load hisat2/2.2.1
hisat2-build -p 4 --exon genome.exon --ss genome.ss genome.fa genome_ath_transStart doing alignment and read counting.
module load hisat2/2.2.1
module load samtools/intel/1.12
module load subread/intel/2.0.1
hisat2 -p 2 -x /scratch/jh6577/tutorial_workshop/genome/genome_ath_trans -U DIV1_p1_clean.fq.gz -S DIV1_p1_clean.sam 2> info.txt
REF="/scratch/jh6577/tutorial_workshop/genome/genome_ath_trans"
hisat2 -p 2 -x ${REF} -U DIV1_p1_clean.fq.gz 2>DIV1_p1.aln.info | samtools view -bSh - | samtools sort -o DIV1_p1.bam -
samtools view -h DIV1_p1.bam|head -n 20
hisat2 -p 2 -x ${REF} -U EV_p1_clean.fq.gz 2>EV_p1.aln.info | samtools view -bSh - | samtools sort -o EV_p1.bam -
GTF="/scratch/jh6577/tutorial_workshop/genome/genome.gtf"
featureCounts -T 2 -a "${GTF}" -o final.featureCounts *.bamPut everything in a loop in Linux.
for file in *.fq.gz; do
output="${file%.fq.gz}_clean.fq.gz"
fastp -l 20 --thread 1 -y -t 1 -x -a AGATCGGAAGAGC -f 2 -i "$file" -o "$output"
done
REF="/scratch/jh6577/tutorial_workshop/genome/genome_ath_trans"
for file in *_clean.fq.gz; do
output_bam="${file%.fq.gz}.bam"
output_info="${file%.fq.gz}.aln.info"
hisat2 -p 2 -x ${REF} -U "$file" 2>"$output_info" | samtools view -bSh - | samtools sort -o "$output_bam" -
donefastp_array_job.slurm
#!/bin/bash
#SBATCH --job-name=fastp_array # Job name
#SBATCH --array=1-2 # Number of *.fq.gz files
#SBATCH --output=fastp_%A_%a.out # Standard output and error log (%A for array job ID, %a for array index)
#SBATCH --ntasks=1 # Number of CPU cores per task
#SBATCH --time=00:10:00 # Time limit hrs:min:sec
#SBATCH --mem=2gb # Job memory request
# Get file based on array task ID
FILE=$(ls *.fq.gz | sed -n "${SLURM_ARRAY_TASK_ID}p")
# Create output filename
OUTPUT="${FILE%.fq.gz}_clean.fq.gz"
# Run fastp
fastp -l 20 --thread 1 -y -t 1 -x -a AGATCGGAAGAGC -f 2 -i "$FILE" -o "$OUTPUT"Workflows:
Once you have the count matrix, you can use R to analyze it. DESeq2, limma, edgeR are the most popular packages for Differential Expression analysis.