Neural network-based integration of polygenic and clinical information: Development and validation of a prediction model for 10 year risk of major adverse cardiac events in the UK Biobank cohort
Code related to the paper "Neural network-based integration of polygenic and clinical information: Development and validation of a prediction model for 10 year risk of major adverse cardiac events in the UK Biobank cohort". This repo is a python package for preprocessing UK Biobank data and preprocessing, training and evaluating the NeuralCVD score.
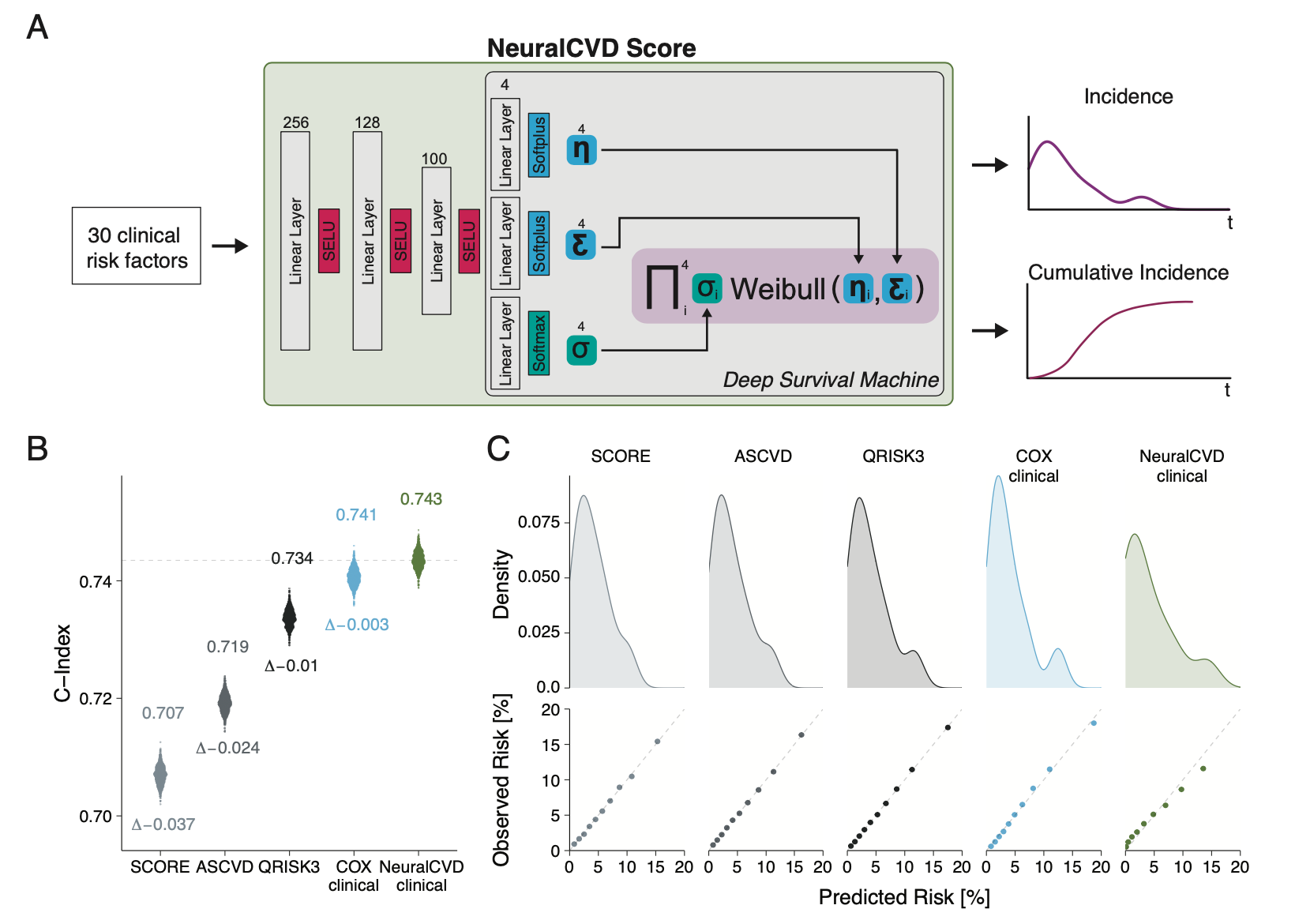
NeuralCVD is based on the fantastic Deep Survival Machines Paper, the original implementation can be found here.
This repo contains code to preprocess UK Biobank data, train the NeuralCVD score and analyze/evaluate its performance.
- Preprocessing involves: parsing primary care records for desired diagnosis, aggregating the cardiovascular risk factors analyzed in the study and calculating predefined polygenic risk scores.
- Training involves Model specification via pytorch-lightning and hydra.
- Postprocessing involve extensive benchmarks with linear Models, and calculation of bootstrapped metrics.
- First, install dependencies
# clone project
git clone https://github.com/thbuerg/NeuralCVD
# install project
cd NeuralCVD
pip install -e .
pip install -r requirements.txt-
Download UK Biobank data. Execute preprocessing notebooks on the downloaded data.
-
Edit the
.yamlconfig files inneuralcvd/experiments/config/:
setup:
project_name: <YourNeptuneSpace>/<YourProject>
root_dir: absolute/path/to/this/repo/
experiment:
tabular_filepath: path/to/processed/data-
Set up Neptune.ai
-
Train the NeuralCVD Model (make sure you are on a machine w/ GPU)
# module folder
cd neuralcvd
# run training
bash experiments/run_NeuralCVD_S.sh@article{steinfeldt2022neural,
title={Neural network-based integration of polygenic and clinical information: development and validation of a prediction model for 10-year risk of major adverse cardiac events in the UK Biobank cohort},
author={Steinfeldt, Jakob and Buergel, Thore and Loock, Lukas and Kittner, Paul and Ruyoga, Greg and zu Belzen, Julius Upmeier and Sasse, Simon and Strangalies, Henrik and Christmann, Lara and Hollmann, Noah and others},
journal={The Lancet Digital Health},
volume={4},
number={2},
pages={e84--e94},
year={2022},
publisher={Elsevier}
}