Quickly produce pretty dotplots from minimap mappings using R/ggplot2.
- R packages: ggplot2, stringr, scales, argparse
- curl, gzip (for samples only)
git clone https://github.com/thackl/minidot.git
make sample-prochlorococcus make sample-arabidopsis
Make sample commands will download required data, set up minimap (if not already in $PATH) and run minidot.
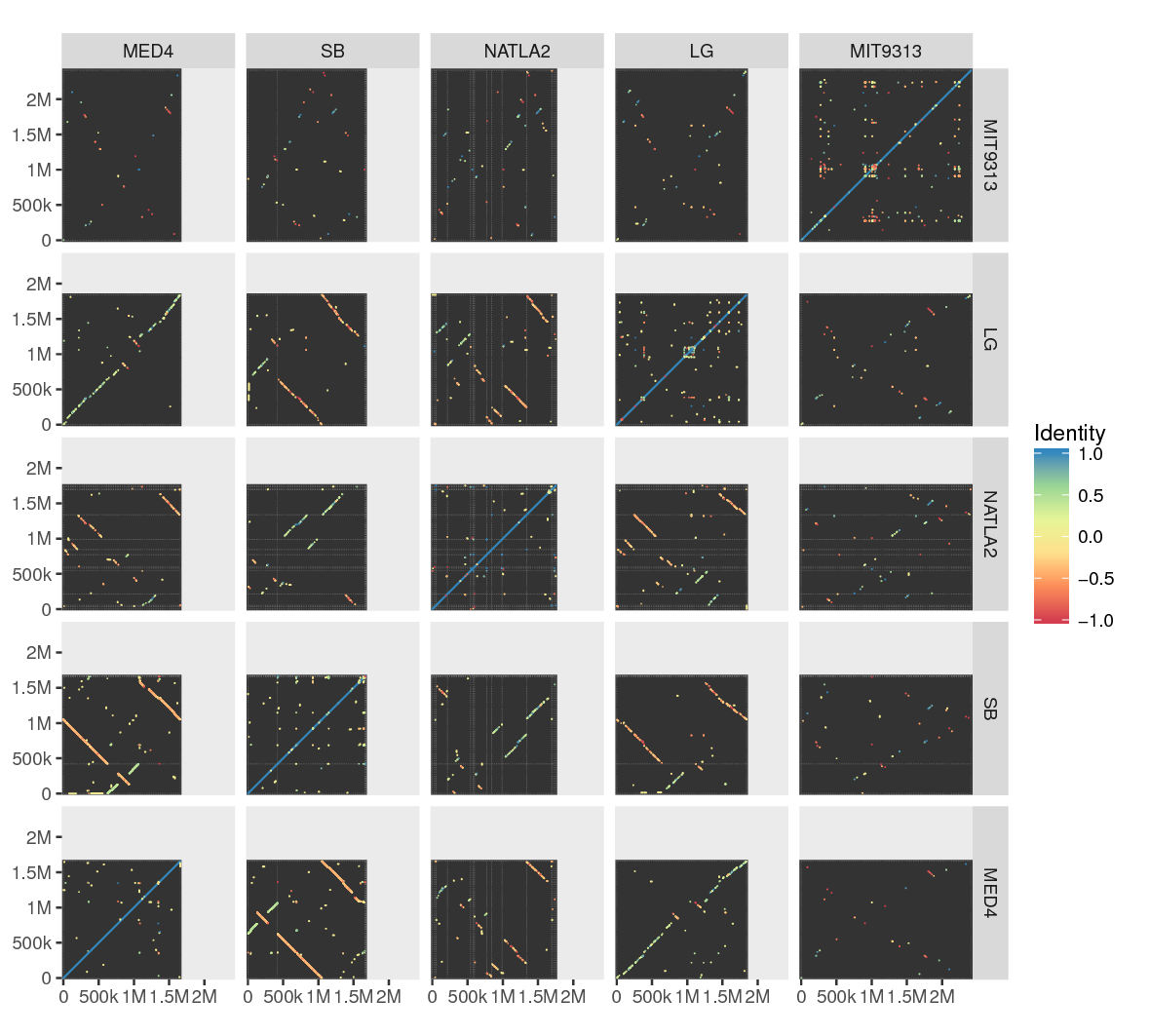
# actual minidot command invoked by make sample-prochlorococcus after setting up the data minidot -s -o prochlorococcus.pdf MED4.da SB.fa NATLA2.fa LG.fa MIT9313.fa
Quickly compare genome organization among multiple bacterial genomes (finished and draft).
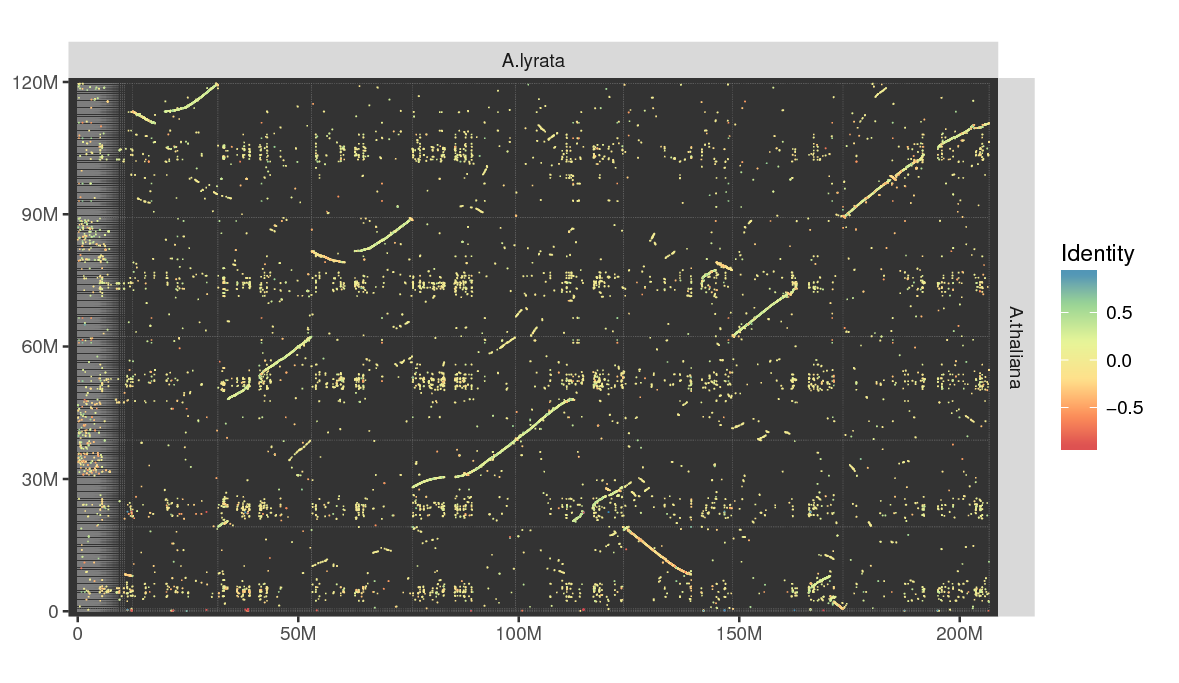
# actual minidot command invoked by make sample-arabidopsis after setting up the data minidot -o arabidopsis.pdf A.lyrata.fa A.thaliana.fa
Compare two related eukaryotic genomes. Larger genomes are automatically run in
fast mode, which is less sensitive and looks only for larger stretches of
similarity (>500bp).
For A.t. vs A.l., mapping & plotting takes about 1.5 minutes (desktop PC, 3 threads, 8GB RAM). That’s probably faster than downloading the genomes in the first place…
Any kind of feedback is highly appreciated. Feel free to either report issues directly via github or shoot me an email.