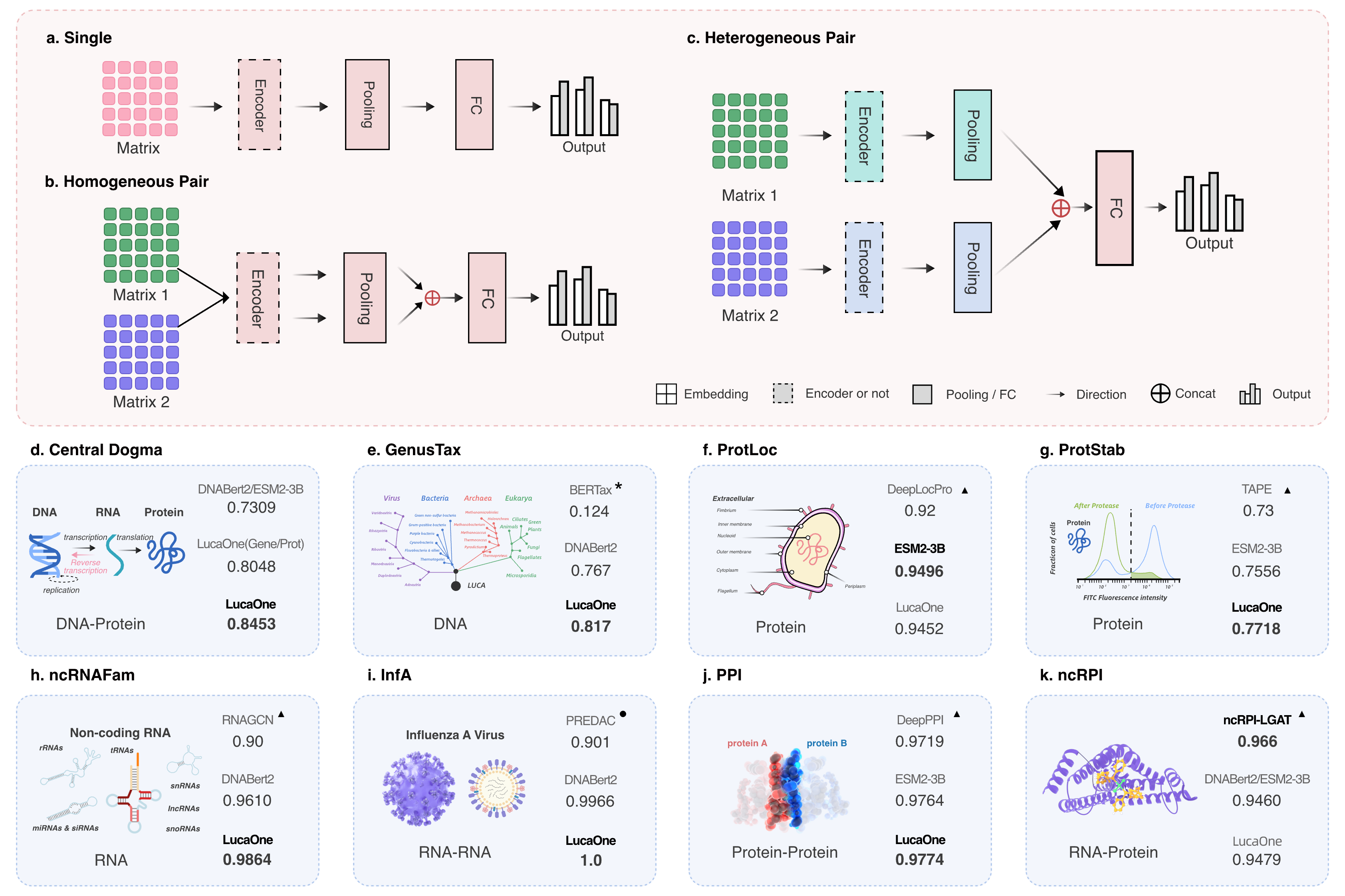
Three distinct networks correspond to three different types of inputs:
- LucaBase(Single)
- LucaPPI(Homogeneous Pair)
- LucaPPI2(Heterogeneous Pair)
Fig. 1 Downstream task network with three input types and results comparison of 8 ver- ification tasks.
-
Central Dogma(Central Dogma of Molecular Biology)
Input: DNA + Protein(heterogeneous double sequence)
Network: LucaPPI2(src/ppi/models/LucaPPI2) -
SupKTax(Genus Taxonomy Annotation)
Input: DNA(single sequence)
Network: LucaBase(src/common/luca_base) -
GenusTax(SuperKingdom Taxonomy Annotation)
Input: DNA(single sequence)
Network: LucaBase(src/common/luca_base) -
SpeciesTax(Species Taxonomy Annotation)
Input: DNA(single sequence)
Network: LucaBase(src/common/luca_base) -
ProtLoc(Prokaryotic Protein Subcellular Location)
Input: Protein(single sequence)
Network: LucaBase(src/common/luca_base) -
ProtStab(Protein Stability)
Input: Protein(single sequence)
Network: LucaBase(src/common/luca_base) -
ncRNAFam(Non-coding RNA Family)
Input: RNA(single sequence)
Network: LucaBase(src/common/luca_base) -
InfA(Influenza A Antigenic Relationship Prediction)
Input: RNA + RNA(homogeneous double sequence)
Network: LucaPPI(src/ppi/models/LucaPPI) -
PPI(Protein-Protein Interaction)
Input: Protein + Protein(homogeneous double sequence)
Network: LucaPPI(src/ppi/models/LucaPPI) -
ncRPI(ncRNA-Protein Interactions)
Input: DNA + Protein(heterogeneous double sequence)
Network: LucaPPI2(src/ppi/models/LucaPPI2)
sudo yum update
sudo yum install git-all
sudo apt-get update
sudo apt install git-all
wget https://repo.anaconda.com/archive/Anaconda3-2022.05-Linux-x86_64.sh
sh Anaconda3-2022.05-Linux-x86_64.sh
source ~/.bashrc
conda create -n lucaone_tasks python=3.9.13
conda activate lucaone_tasks
pip install -r requirements.txt -i https://pypi.tuna.tsinghua.edu.cn/simple
Downstream Tasks Dataset FTP: Dataset for LucaOneTasks
Copy the 10 datasets from http://47.93.21.181/lucaone/DownstreamTasksDataset/dataset/* into the directory ./dataset/
Trained LucaOne Checkpoint FTP: TrainedCheckPoint for LucaOne
Notice
The project will download automatically LucaOne Trained-CheckPoint from FTP.
When downloading automatically failed, you can manually download:
Copy the TrainedCheckPoint Files(models/ + logs/) from http://47.93.21.181/lucaone/TrainedCheckPoint/* into the directory ./llm/
Methods of using embedding:
In this project, the sequence is embedded during the training downstream task(./src/encoder.py).
We can also embed the dataset and store into a predefined folder, then build and train the downstream network.
the script of embedding a dataset(./src/llm/lucagplm/get_embedding.py):
Notice:
a. need to specify the column index of the sequence id(id_idx*) and sequence(seq_idx), starting index: 0.
b. The sequence id must be globally unique in the input file and cannot contain special characters (because the embedding file stored is named by the sequence id).
# for protein
cd ./src/llm/lucagplm
export CUDA_VISIBLE_DEVICES="0,1,2,3,4,5,6,7,8"
python get_embedding.py \
--llm_dir ../../.. \
--llm_type lucaone_gplm \
--llm_version v2.0 \
--llm_task_level token_level,span_level,seq_level,structure_level \
--llm_time_str 20231125113045 \
--llm_step 5600000 \
--embedding_type matrix \
--trunc_type right \
--truncation_seq_length 100000 \
--matrix_add_special_token \
--seq_type prot \
--input_file ../../../data/test_data/prot/test_prot.csv \
--id_idx 2 \
--seq_idx 3 \
--save_path ../../../embedding/lucaone/test_data/prot/test_prot \
--embedding_complete \
--embedding_complete_seg_overlap \
--gpu 0
# for DNA or RNA
cd ./src/llm/lucagplm
export CUDA_VISIBLE_DEVICES="0,1,2,3,4,5,6,7,8"
python get_embedding.py \
--llm_dir ../../.. \
--llm_type lucaone_gplm \
--llm_version v2.0 \
--llm_task_level token_level,span_level,seq_level,structure_level \
--llm_time_str 20231125113045 \
--llm_step 5600000 \
--embedding_type matrix \
--trunc_type right \
--truncation_seq_length 100000 \
--matrix_add_special_token \
--seq_type gene \
--input_file ../../../data/test_data/gene/test_gene.csv \
--id_idx 0 \
--seq_idx 1 \
--save_path ../../../embedding/lucaone/test_data/gene/test_gene \
--embedding_complete \
--embedding_complete_seg_overlap \
--gpu 0 Notice:
a. The sequence id must be globally unique in the input file and cannot contain special characters (because the embedding file stored is named by the sequence id).
# for protein
cd ./src/llm/lucagplm
export CUDA_VISIBLE_DEVICES="0,1,2,3,4,5,6,7,8"
python get_embedding.py \
--llm_dir ../../.. \
--llm_type lucaone_gplm \
--llm_version v2.0 \
--llm_task_level token_level,span_level,seq_level,structure_level \
--llm_time_str 20231125113045 \
--llm_step 5600000 \
--embedding_type matrix \
--trunc_type right \
--truncation_seq_length 100000 \
--matrix_add_special_token \
--seq_type prot \
--input_file ../../../data/test_data/prot/test_prot.fasta \
--save_path ../../../embedding/lucaone/test_data/prot/test_prot \
--embedding_complete \
--embedding_complete_seg_overlap \
--gpu 0 # for DNA or RNA
cd ./src/llm/lucagplm
export CUDA_VISIBLE_DEVICES="0,1,2,3,4,5,6,7,8"
python get_embedding.py \
--llm_dir ../../.. \
--llm_type lucaone_gplm \
--llm_version v2.0 \
--llm_task_level token_level,span_level,seq_level,structure_level \
--llm_time_str 20231125113045 \
--llm_step 5600000 \
--embedding_type matrix \
--trunc_type right \
--truncation_seq_length 100000 \
--matrix_add_special_token \
--seq_type gene \
--input_file ../../../data/test_data/gene/test_gene.fasta \
--save_path ../../../embedding/lucaone/test_data/gene/test_gene \
--embedding_complete \
--embedding_complete_seg_overlap \
--gpu 0 -
LucaOne checkpoint parameters:
- llm_dir: the path for storing the checkpoint LucaOne model,default:
../../../ - llm_type: the type of LucaOne, default: lucagplm
- llm_version: the version of LucaOne, default: v2.0
- llm_task_level: the pretrained tasks of LucaOne, default: token_level,span_level,seq_level,structure_level
- llm_time_str: the trained time str of LucaOne, default: 20231125113045
- llm_step: the trained checkpoint of LucaOne, default: 5600000
- llm_dir: the path for storing the checkpoint LucaOne model,default:
-
Important parameters:
- embedding_type:
matrixorvector, output the embedding matrix or [CLS] vector for the entire sequence, recommend: matrix. - trunc_type: truncation type:
rightorleft, truncation when the sequence exceeds the maximum length. - truncation_seq_length: the maximum length for embedding(not including [CLS] and [SEP]), itself does not limit the length, depending on the capacity of GPU.
- matrix_add_special_token: if the embedding is matrix, whether the matrix includes [CLS] and [SEP] vectors.
- seq_type: type of input sequence:
geneorprot,genefor nucleic acid(DNA or RNA),protfor protein. - input_file: the input file path for embedding(format: csv or fasta). The seq_id in the file must be unique and cannot contain special characters.
- save_path: the saving dir for storing the embedding file.
- embedding_complete: When
embedding_completeis set,truncation_seq_lengthis invalid. If the GPU memory is not enough to infer the entire sequence at once, it is used to determine whether to perform segmented completion (if this parameter is not used, 0.95*len is truncated each time until the CPU can process the length). - embedding_complete_seg_overlap: When
embedding_completeis set, whether the method of overlap is applicable to segmentation(overlap sliding window) - gpu: the gpu id to use(-1 for cpu).
- embedding_type:
-
Optional parameters:
- id_idx & seq_idx: when the input file format is csv file, need to use
id_idxandseq_idxto specify the column index in the csv (starting with 0).
- id_idx & seq_idx: when the input file format is csv file, need to use
The running scripts of 10 downstream tasks in three directories:
-
src/training/lucaone:
The running scripts of the 10 downstream tasks were based on LucaOne's embedding (Fig. 4 in our paper). -
src/training/downstream_tasks:
A complete comparison on the 10 downstream tasks.
These comparisons were based on the embedding of LucaOne, DNABert2, and ESM2-3B. (Table.S5 in our paper). -
src/training/lucaone_separated:
The task script with the embedding based on the LucaOne of separated nucleic acid and protein training(LucaOne-Gene/LucaOne-Prot). (Fig. 3 in our paper).
FTP:
Pre-training data, code, and trained checkpoint of LucaOne, embedding inference code, downstream validation tasks data & code, and other materials are available: FTP.
Details:
The LucaOne's model code is available at: LucaOne Github or LucaOne.
The trained-checkpoint files are available at: TrainedCheckPoint.
LucaOne's representational inference code is available at: LucaOneApp Github or LucaOneApp.
The project of 8 downstream tasks is available at: LucaOneTasks Github or LucaOneTasks.
The pre-training dataset of LucaOne is opened at: PreTrainingDataset.
The datasets of downstream tasks are available at: DownstreamTasksDataset .
Other supplementary materials are available at: Others .
Yong He, Zhaorong Li, Pan Fang
@article {LucaOne,
author = {Yong He and Pan Fang and Yongtao Shan and Yuanfei Pan and Yanhong Wei and Yichang Chen and Yihao Chen and Yi Liu and Zhenyu Zeng and Zhan Zhou and Feng Zhu and Edward C. Holmes and Jieping Ye and Jun Li and Yuelong Shu and Mang Shi and Zhaorong Li},
title = {LucaOne: Generalized Biological Foundation Model with Unified Nucleic Acid and Protein Language},
elocation-id = {2024.05.10.592927},
year = {2024},
doi = {10.1101/2024.05.10.592927},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/05/14/2024.05.10.592927},
eprint = {https://www.biorxiv.org/content/early/2024/05/14/2024.05.10.592927.full.pdf},
journal = {bioRxiv}
}