This package is under development.
This package estimates the time-varying reproduction number, rate of
spread, and doubling time using a range of open-source tools and current
best practices. It aims to help users avoid some of the limitations of
naive implementations in a framework that is informed by community
feedback and is under active development. It estimates the time-varying
reproduction number on cases by date of infection (using a similar
approach to that implemented in the
{EpiEstim}). Imputed
infections are then mapped to observed data (for example cases by date
of report) via a series of uncertain delay distributions (in this
example an incubation period and a reporting delay) and a reporting
model that can include weekly periodicity. The default model uses a
non-stationary Gaussian process to estimate the time-varying
reproduction number but optionally a stationary Gaussian process may be
used (faster to estimate but reduced performance for real time
estimates) and arbitrary breakpoints can be defined. Propagating
uncertainty from all inputs into the final parameter estimates (helping
to mitigate spurious findings) is handled internally. Time-varying
estimates of the rate of growth are derived from the time-varying
reproduction estimates and the uncertain generation time. Optionally,
the time-varying reproduction number can be forecast forwards in time
using an integration with the
{EpiSoon} package and converted to
a case forecast using the renewal equation. Alternatively, the
time-varying reproduction number and cases can be forecast using a
Gaussian process. As a standalone tool non-parametric back-calculation
is also supported using a novel formulation based on a smoothed mean
delay shift of reported cases combined with a Gaussian process to
determine the most likely outbreak trajectory.
Install the stable version of the package using
{drat}:
install.packages("drat")
drat:::add("epiforecasts")
install.packages("EpiNow2")Install the development version of the package with:
remotes::install_github("epiforecasts/EpiNow2")Windows users will need a working installation of Rtools in order to build the package from source. See here for a guide to installing Rtools for use with Stan (which is the statistical modeling platform used for the underlying model). For simple deployment/development a prebuilt docker image is also available (see documentation here).
{EpiNow} is designed to be used with a single function call or to be
used in an ad-hoc fashion via individual function calls. In the
following section we give an overview of the simple use case. For more
on using each function see the function
documentation.
The core functions are:
epinow,
regional_epinow,
estimate_infections,
and
forecast_infections.
estimate_infections
can be use on its own to infer the underlying infection case curve from
reported cases with Rt optionally returned (on by default). Estimating
the underlying infection case curve alone is substantially less
computationally demanding than also estimating Rt.
Distributions can either be fitted using package functionality or
determined elsewhere and then defined with uncertainty for use in
{EpiNow2}. When data is supplied a subsampled bootstrapped lognormal
will be fit (to account for uncertainty in the observed data without
being biased by changes in incidence). An arbitrary number of delay
distributions are supported with the most common use case likely to be a
incubation period followed by a reporting delay.
reporting_delay <- EpiNow2::bootstrapped_dist_fit(rlnorm(100, log(6), 1))
## Set max allowed delay to 30 days to truncate computation
reporting_delay$max <- 30
reporting_delay
#> $mean
#> [1] 1.698169
#>
#> $mean_sd
#> [1] 0.1546306
#>
#> $sd
#> [1] 1.114934
#>
#> $sd_sd
#> [1] 0.1117431
#>
#> $max
#> [1] 30Here we define the incubation period and generation time based on literature estimates for Covid-19 (see here for the code that generates these estimates).
generation_time <- list(mean = EpiNow2::covid_generation_times[1, ]$mean,
mean_sd = EpiNow2::covid_generation_times[1, ]$mean_sd,
sd = EpiNow2::covid_generation_times[1, ]$sd,
sd_sd = EpiNow2::covid_generation_times[1, ]$sd_sd,
max = 30)
incubation_period <- list(mean = EpiNow2::covid_incubation_period[1, ]$mean,
mean_sd = EpiNow2::covid_incubation_period[1, ]$mean_sd,
sd = EpiNow2::covid_incubation_period[1, ]$sd,
sd_sd = EpiNow2::covid_incubation_period[1, ]$sd_sd,
max = 30)This function represents the core functionality of the package and
includes results reporting, plotting and optional saving. It requires a
data frame of cases by date of report and the distributions defined
above. An additional forecasting module is supported via EpiSoon and
companion packages (see documentation for an example).
Load example case data from {EpiNow2}.
reported_cases <- EpiNow2::example_confirmed[1:50]
head(reported_cases)
#> date confirm
#> 1: 2020-02-22 14
#> 2: 2020-02-23 62
#> 3: 2020-02-24 53
#> 4: 2020-02-25 97
#> 5: 2020-02-26 93
#> 6: 2020-02-27 78Estimate cases by date of infection, the time-varying reproduction
number, the rate of growth and forecast these estimates into the future
by 7 days. Summarise the posterior and return a summary table and plots
for reporting purposes. If a target_folder is supplied results can be
internally saved (with the option to also turn off explicit returning of
results). Note that for real use cases more samples and a longer warm
up may be needed.
estimates <- EpiNow2::epinow(reported_cases = reported_cases,
generation_time = generation_time,
delays = list(incubation_period, reporting_delay),
horizon = 7, samples = 1000, warmup = 200,
cores = 4, chains = 4, verbose = TRUE,
adapt_delta = 0.95)
#> [[1]]
#> Stan model 'estimate_infections' does not contain samples.
#>
#> [[2]]
#> Stan model 'estimate_infections' does not contain samples.
names(estimates)
#> [1] "estimates" "estimated_reported_cases"
#> [3] "summary" "plots"Both summary measures and posterior samples are returned for all parameters in an easily explored format.
estimates$estimates
#> $samples
#> variable parameter time date sample value
#> 1: infections imputed_infections 1 2020-02-07 1 3.000
#> 2: infections imputed_infections 2 2020-02-08 1 16.000
#> 3: infections imputed_infections 3 2020-02-09 1 22.000
#> 4: infections imputed_infections 4 2020-02-10 1 37.000
#> 5: infections imputed_infections 5 2020-02-11 1 41.000
#> ---
#> 128068: prior_infections prior_infections 68 2020-04-14 1 2581.666
#> 128069: prior_infections prior_infections 69 2020-04-15 1 2517.820
#> 128070: prior_infections prior_infections 70 2020-04-16 1 2455.553
#> 128071: prior_infections prior_infections 71 2020-04-17 1 2394.826
#> 128072: prior_infections prior_infections 72 2020-04-18 1 2335.601
#> strat type
#> 1: <NA> estimate
#> 2: <NA> estimate
#> 3: <NA> estimate
#> 4: <NA> estimate
#> 5: <NA> estimate
#> ---
#> 128068: <NA> forecast
#> 128069: <NA> forecast
#> 128070: <NA> forecast
#> 128071: <NA> forecast
#> 128072: <NA> forecast
#>
#> $summarised
#> date variable strat type bottom top
#> 1: 2020-02-22 R <NA> estimate 0.7564282 1.414028
#> 2: 2020-02-23 R <NA> estimate 0.8846257 1.448911
#> 3: 2020-02-24 R <NA> estimate 0.9631228 1.437619
#> 4: 2020-02-25 R <NA> estimate 1.0954422 1.468108
#> 5: 2020-02-26 R <NA> estimate 1.2025306 1.512359
#> ---
#> 324: 2020-04-14 reported_cases <NA> forecast 374.0000000 6874.000000
#> 325: 2020-04-15 reported_cases <NA> forecast 142.0000000 5239.000000
#> 326: 2020-04-16 reported_cases <NA> forecast 361.0000000 6498.000000
#> 327: 2020-04-17 reported_cases <NA> forecast 458.0000000 9039.000000
#> 328: 2020-04-18 reported_cases <NA> forecast 204.0000000 7451.000000
#> lower upper median mean sd
#> 1: 0.9459273 1.234976 1.105647 1.113589 2.057302e-01
#> 2: 0.9905997 1.236385 1.162262 1.166212 1.754555e-01
#> 3: 1.1198978 1.322856 1.228027 1.226912 1.457745e-01
#> 4: 1.2053574 1.371572 1.295255 1.294681 1.183796e-01
#> 5: 1.3104278 1.437038 1.368334 1.367978 9.674816e-02
#> ---
#> 324: 1075.0000000 2992.000000 2746.500000 3550.534000 3.046264e+03
#> 325: 1057.0000000 2669.000000 2226.000000 2855.966000 2.525423e+03
#> 326: 653.0000000 2748.000000 2574.500000 3628.484000 4.226875e+03
#> 327: 1045.0000000 3289.000000 2876.000000 4804.820000 8.242916e+03
#> 328: 917.0000000 2695.000000 2378.500000 4145.540000 8.458841e+03Reported cases are returned separately in order to ease reporting of forecasts and model evaluation.
estimates$estimated_reported_cases
#> $samples
#> date sample cases type
#> 1: 2020-02-22 1 119 gp_rt
#> 2: 2020-02-23 1 97 gp_rt
#> 3: 2020-02-24 1 73 gp_rt
#> 4: 2020-02-25 1 331 gp_rt
#> 5: 2020-02-26 1 210 gp_rt
#> ---
#> 28496: 2020-04-14 500 3076 gp_rt
#> 28497: 2020-04-15 500 1456 gp_rt
#> 28498: 2020-04-16 500 3279 gp_rt
#> 28499: 2020-04-17 500 5650 gp_rt
#> 28500: 2020-04-18 500 2047 gp_rt
#>
#> $summarised
#> date type bottom top lower upper median mean sd
#> 1: 2020-02-22 gp_rt 31 230 66 140 121.0 135.552 69.91704
#> 2: 2020-02-23 gp_rt 43 306 83 188 166.5 174.868 86.47373
#> 3: 2020-02-24 gp_rt 47 357 117 235 185.0 206.276 112.55849
#> 4: 2020-02-25 gp_rt 36 363 113 229 194.0 215.566 113.04736
#> 5: 2020-02-26 gp_rt 60 354 106 215 186.5 202.662 97.72685
#> 6: 2020-02-27 gp_rt 50 446 115 263 229.5 260.560 136.64177
#> 7: 2020-02-28 gp_rt 93 632 159 366 326.5 357.236 184.91553
#> 8: 2020-02-29 gp_rt 93 570 157 332 289.0 329.884 179.01133
#> 9: 2020-03-01 gp_rt 99 692 184 407 356.5 398.796 206.27769
#> 10: 2020-03-02 gp_rt 129 765 184 410 384.0 431.012 218.11775
#> 11: 2020-03-03 gp_rt 100 777 177 433 401.5 440.188 228.74989
#> 12: 2020-03-04 gp_rt 82 711 231 483 383.5 428.928 232.34748
#> 13: 2020-03-05 gp_rt 110 1073 256 638 557.5 627.182 337.31880
#> 14: 2020-03-06 gp_rt 221 1420 463 946 773.0 862.396 454.21571
#> 15: 2020-03-07 gp_rt 172 1495 383 883 748.5 852.600 458.96804
#> 16: 2020-03-08 gp_rt 208 2034 539 1208 1051.0 1170.814 633.95379
#> 17: 2020-03-09 gp_rt 320 2356 554 1357 1224.5 1371.076 748.93441
#> 18: 2020-03-10 gp_rt 389 2480 732 1567 1292.0 1393.288 695.64383
#> 19: 2020-03-11 gp_rt 320 2524 572 1484 1274.5 1452.322 800.00725
#> 20: 2020-03-12 gp_rt 616 3651 1010 2093 1828.0 2071.244 1078.45410
#> 21: 2020-03-13 gp_rt 614 5053 1234 2818 2522.5 2790.782 1596.56347
#> 22: 2020-03-14 gp_rt 527 4576 1396 2896 2467.0 2762.042 1438.36390
#> 23: 2020-03-15 gp_rt 871 5761 1820 3758 3133.5 3397.316 1670.04978
#> 24: 2020-03-16 gp_rt 858 6667 1576 3934 3475.0 3845.562 2081.31864
#> 25: 2020-03-17 gp_rt 706 6464 1596 3892 3478.0 3787.802 1993.28062
#> 26: 2020-03-18 gp_rt 778 6016 1930 3931 3289.0 3619.738 1999.37828
#> 27: 2020-03-19 gp_rt 1077 8256 2289 4937 4260.0 4733.790 2656.64362
#> 28: 2020-03-20 gp_rt 1540 10942 2777 6104 5412.0 6057.274 3536.53122
#> 29: 2020-03-21 gp_rt 1546 8833 2626 5686 4685.5 5106.524 2473.41052
#> 30: 2020-03-22 gp_rt 1587 10402 3436 7084 5589.5 6105.698 3008.77807
#> 31: 2020-03-23 gp_rt 1957 10848 2963 6316 5578.0 6159.720 3310.74146
#> 32: 2020-03-24 gp_rt 1407 9299 3098 6177 4864.5 5237.604 2629.14262
#> 33: 2020-03-25 gp_rt 1180 7753 2655 5223 4071.5 4531.636 2244.29211
#> 34: 2020-03-26 gp_rt 1459 9687 2813 6137 5298.5 5660.282 2881.43164
#> 35: 2020-03-27 gp_rt 1124 11926 3790 7785 6376.5 7062.978 3782.16643
#> 36: 2020-03-28 gp_rt 1332 10161 2750 5885 5126.0 5791.106 3224.55387
#> 37: 2020-03-29 gp_rt 1302 10634 2822 6548 5618.0 6133.976 3216.79496
#> 38: 2020-03-30 gp_rt 1824 11113 3573 6901 5301.0 6030.932 3247.57796
#> 39: 2020-03-31 gp_rt 1185 8284 2815 5750 4813.5 5122.896 2435.69280
#> 40: 2020-04-01 gp_rt 1093 7339 2114 4408 3797.5 4226.644 2152.01092
#> 41: 2020-04-02 gp_rt 1307 8866 2482 5361 4540.0 4962.260 2585.04416
#> 42: 2020-04-03 gp_rt 1681 10833 2461 6030 5394.0 5926.578 3038.09044
#> 43: 2020-04-04 gp_rt 895 8175 3056 5857 4380.0 4790.108 2378.61451
#> 44: 2020-04-05 gp_rt 1023 8775 2559 5537 4597.0 5136.340 2794.30555
#> 45: 2020-04-06 gp_rt 1587 8493 1777 4428 4268.0 4857.798 2600.59912
#> 46: 2020-04-07 gp_rt 1023 7298 1742 4217 3668.0 4046.914 2143.49553
#> 47: 2020-04-08 gp_rt 703 5439 1656 3505 3005.5 3254.126 1764.74083
#> 48: 2020-04-09 gp_rt 832 6896 1780 4215 3491.5 3939.996 2226.82248
#> 49: 2020-04-10 gp_rt 564 8140 2065 4793 4016.0 4574.440 2668.13093
#> 50: 2020-04-11 gp_rt 774 6444 1485 3709 3292.5 3714.246 2140.26301
#> 51: 2020-04-12 gp_rt 1033 8003 1624 4166 3636.5 4252.412 2907.12434
#> 52: 2020-04-13 gp_rt 314 7614 1285 3641 3252.5 3945.210 2990.48684
#> 53: 2020-04-14 gp_rt 374 6874 1075 2992 2746.5 3550.534 3046.26356
#> 54: 2020-04-15 gp_rt 142 5239 1057 2669 2226.0 2855.966 2525.42336
#> 55: 2020-04-16 gp_rt 361 6498 653 2748 2574.5 3628.484 4226.87475
#> 56: 2020-04-17 gp_rt 458 9039 1045 3289 2876.0 4804.820 8242.91616
#> 57: 2020-04-18 gp_rt 204 7451 917 2695 2378.5 4145.540 8458.84071
#> date type bottom top lower upper median mean sdA summary table is returned for rapidly understanding the results and for reporting purposes.
estimates$summary
#> measure estimate numeric_estimate
#> 1: New confirmed cases by infection date 2048 (19 -- 7867) <data.table>
#> 2: Expected change in daily cases Unsure 0.72
#> 3: Effective reproduction no. 0.8 (0.2 -- 1.4) <data.table>
#> 4: Rate of growth -0.06 (-0.26 -- 0.11) <data.table>
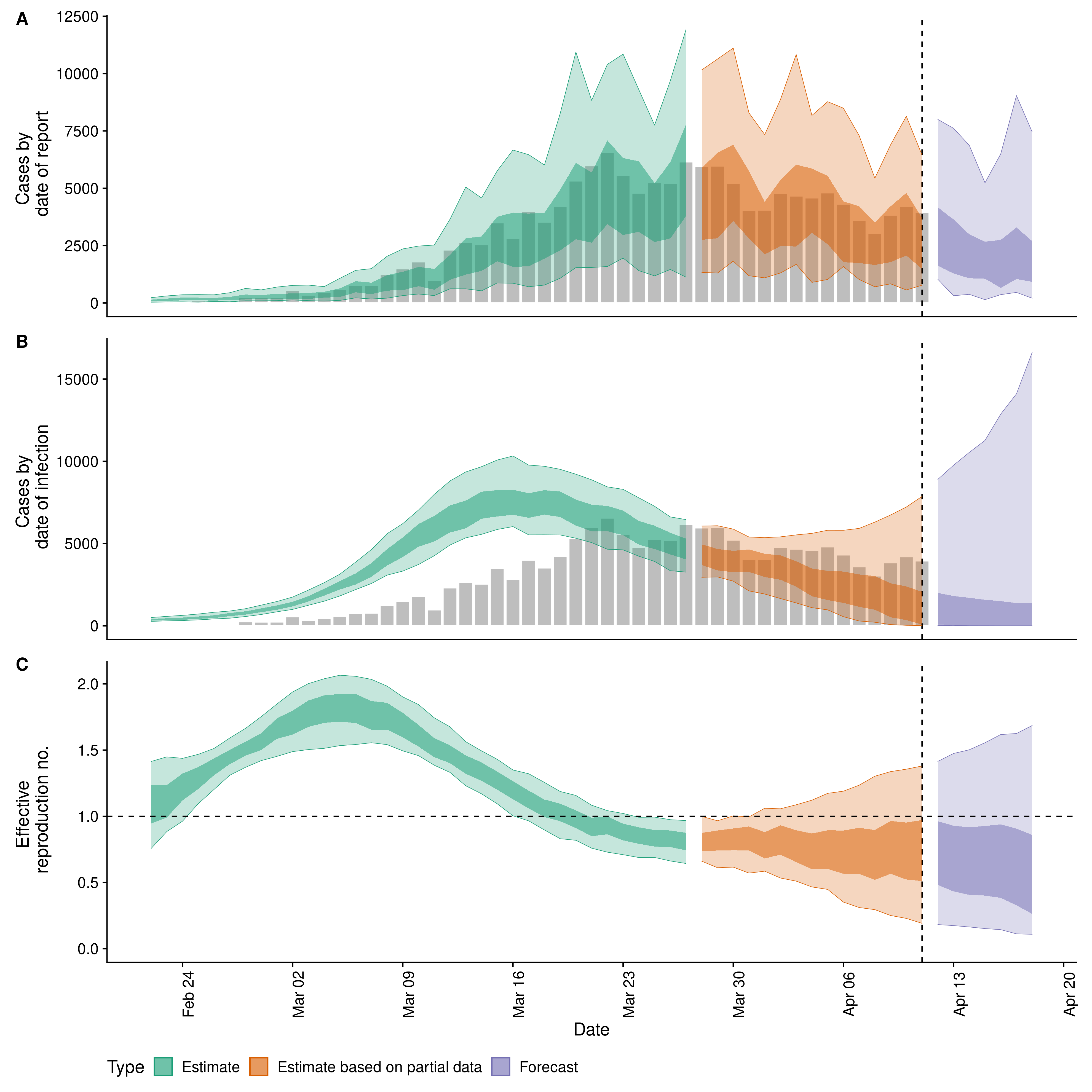
#> 5: Doubling/halving time (days) -10.9 (6.4 -- -2.7) <data.table>A range of plots are returned (with the single summary plot shown below).
estimates$plots$summaryThis function runs the the epinow function across multiple regions in
an efficient manner.
Define cases in multiple regions delineated by the region variable.
reported_cases <- data.table::rbindlist(list(
data.table::copy(reported_cases)[, region := "testland"],
reported_cases[, region := "realland"]))
head(reported_cases)
#> date confirm region
#> 1: 2020-02-22 14 testland
#> 2: 2020-02-23 62 testland
#> 3: 2020-02-24 53 testland
#> 4: 2020-02-25 97 testland
#> 5: 2020-02-26 93 testland
#> 6: 2020-02-27 78 testlandRun the pipeline on each region in turn. The commented code (requires
the {future} package) can be used to run regions in parallel (when in
most scenarios cores should be set to 1).
## future::plan("multisession")
estimates <- EpiNow2::regional_epinow(reported_cases = reported_cases,
generation_time = generation_time,
delays = list(incubation_period, reporting_delay),
horizon = 7, samples = 1000, warmup = 200,
cores = 4, chains = 4, adapt_delta = 0.95,
verbose = TRUE)
#> [[1]]
#> Stan model 'estimate_infections' does not contain samples.
#>
#> [[1]]
#> Stan model 'estimate_infections' does not contain samples.Results from each region are stored in a regional list with across
region summary measures and plots stored in a summary list. All
results can be set to be internally saved by setting the target_folder
and summary_dir arguments.
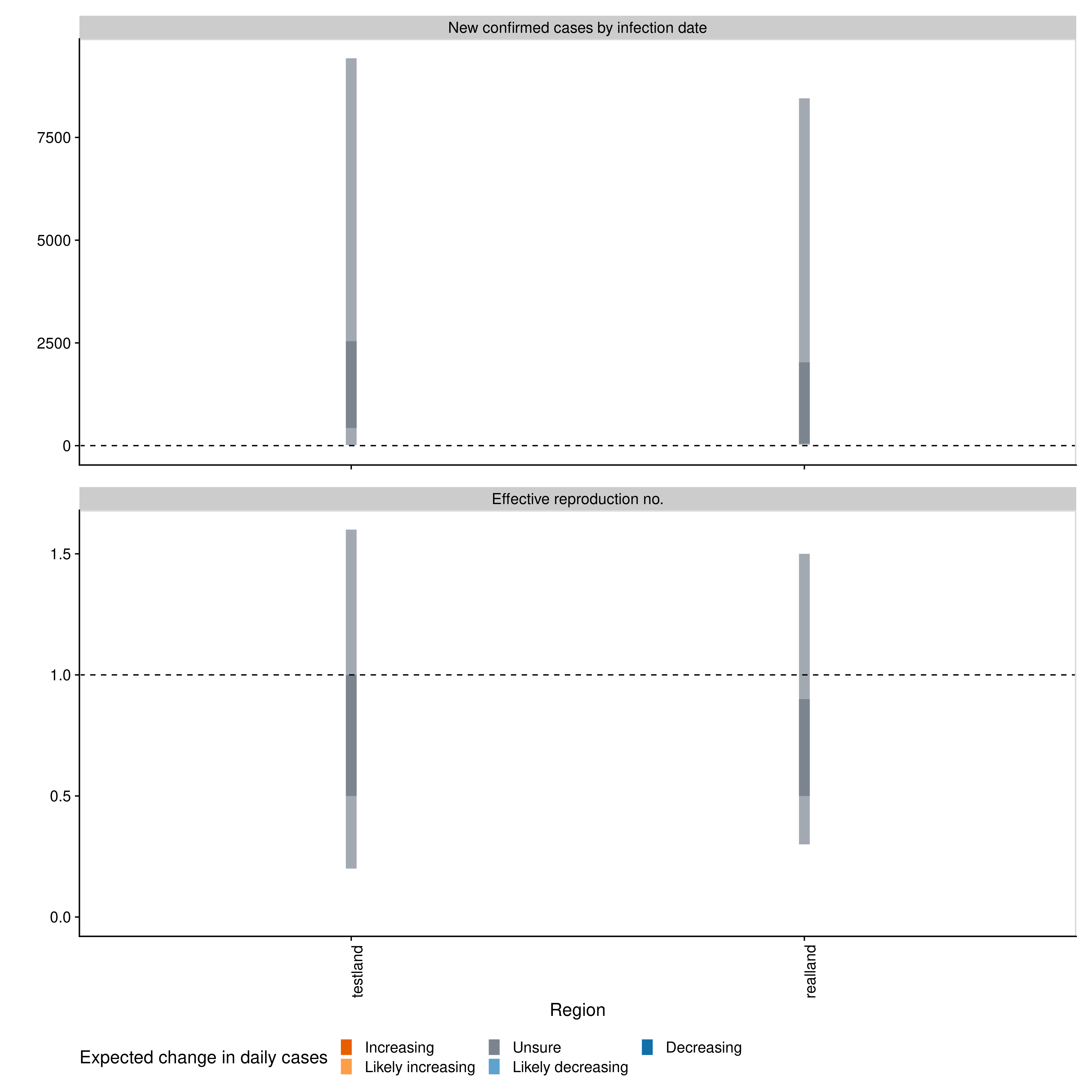
Summary measures that are returned include a table formatted for reporting (along with raw results for further processing).
estimates$summary$summarised_results$table
#> Region New confirmed cases by infection date
#> 1: realland 2012 (26 -- 8451)
#> 2: testland 2190 (13 -- 9426)
#> Expected change in daily cases Effective reproduction no.
#> 1: Unsure 0.8 (0.3 -- 1.5)
#> 2: Unsure 0.8 (0.2 -- 1.6)
#> Rate of growth Doubling/halving time (days)
#> 1: -0.06 (-0.23 -- 0.13) -11.3 (5.2 -- -3)
#> 2: -0.06 (-0.24 -- 0.16) -12.3 (4.4 -- -2.9)A range of plots are again returned (with the single summary plot shown below).
estimates$summary$summary_plotRmarkdown templates are provided in the package (templates) for
semi-automated reporting of estimates. These are currently undocumented
but an example integration can be seen
here.
If using these templates to report your results please highlight our
limitations as these are key to
understanding our results.
File an issue here if you have identified an issue with the package. Please note that due to operational constraints priority will be given to users informing government policy or offering methodological insights. We welcome all contributions, in particular those that improve the approach or the robustness of the code base.