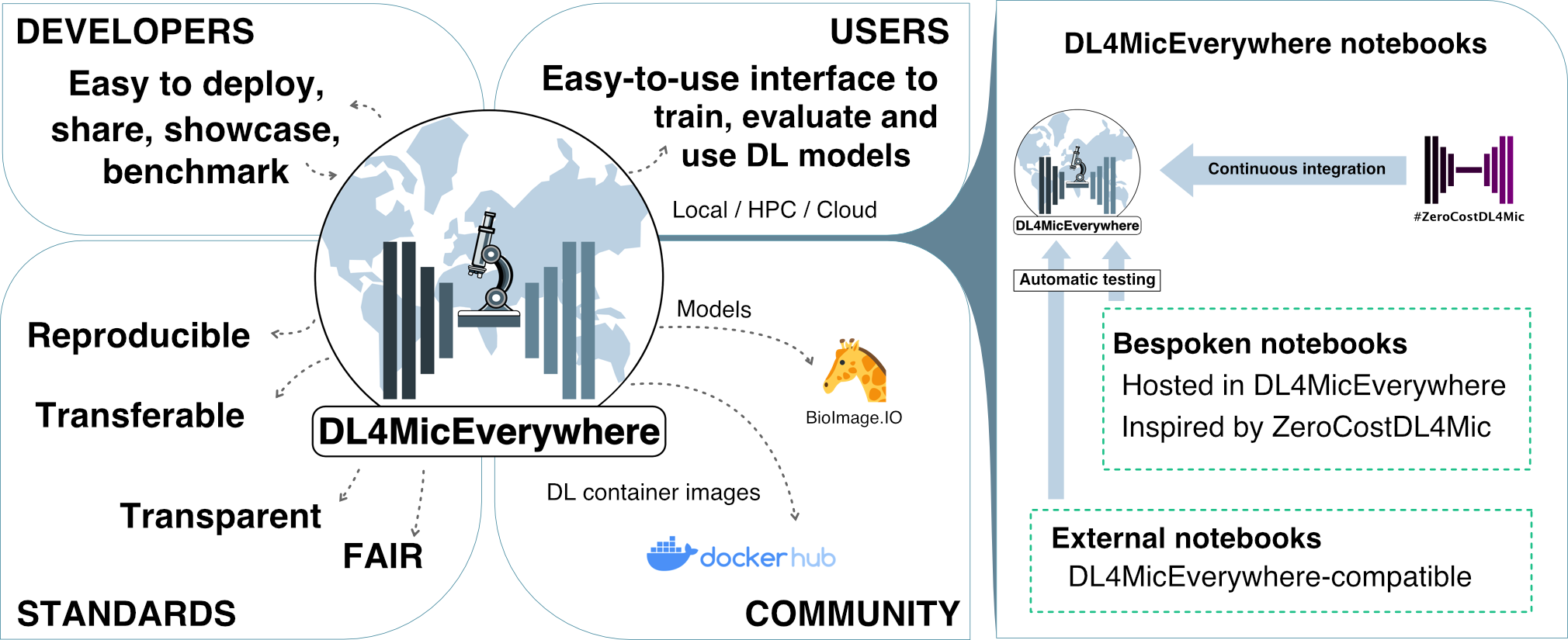
DL4MicEverywhere provides researchers an accessible gateway to state-of-the-art deep learning techniques for bioimage analysis through interactive Jupyter notebooks with intuitive graphical interfaces that require no coding expertise. It uses Docker containers to maximize portability and reproducibility, ensuring seamless operation across diverse computing environments.
DL4MicEverywhere expands the capabilities of ZeroCostDL4Mic by enabling users to run the notebooks locally on their own machines or remotely on diverse computing infrastructure including laptops, workstations, high-performance computing (HPC) and cloud infrastructure. It currently incorporates many existing ZeroCostDL4Mic notebooks for tasks like segmentation, reconstruction, and image translation.
- 15 Jupyter notebooks with intuitive graphical interface requiring no coding (scaling to 28+ soon)
- Docker-based packaging for enhanced portability and reproducibility
- Expanding ZeroCostDL4Mic's capabilities for local use
- Covers a wide range of microscopy analysis tasks, including segmentation, reconstruction, registration, denoising, and more
- Compatible with various computing environments, including laptops, workstations, HPC, and cloud with Docker
- Automated build testing and versioning enhance reliability
- Flexibility: Notebooks can run locally, in the cloud, or on high-performance computing infrastructure. No vendor lock-in.
- Reproducibility: Docker containers encapsulate the full software environment. Explicit versioning maintains stability.
- Transparency: Notebooks and models can be readily shared to enable replication of analyses.
- Accessibility: Interactive widgets and automated build pipelines lower barriers for non-experts.
- Interoperability: Adheres to data standards like BioImage Model Zoo for model sharing.
- Extensibility: Automated testing and Docker building streamlines adding new methods.
DL4MicEverywhere aims to make deep learning more accessible, transparent, and participatory. This enables broader adoption of cutting-edge techniques while enhancing reliability and customization.
- Docker Desktop installed (download)
- For GPU acceleration - NVIDIA GPU + CUDA drivers (setup)
- For the graphical user interface (GUI), Tcl/Tk. (Instructions).
- Clone this repo:
git clone https://github.com/HenriquesLab/DL4MicEverywhere.git - Navigate to the repo directory
- Run
sudo bash launch.shto launch the notebook selection GUI - Choose a notebook and run!
Docker wraps up all dependencies in a tidy bundle. Simply launch and access deep learning workflows through an intuitive interface!
Refer to the User Guide and Installation guidelines for details.
We welcome contributions! Please check out the contributing guidelines to get started.
- User Guide
- Installation
- Data Preparation
- Example Notebooks
- Contributing Guidelines
- Docker desktop
- Technical Design
- Troubleshooting
- Notebook Types
- Notebook List
- FAQ
Let us know if anything needs clarification!
We thank the ZeroCostDL4Mic contributors for their work on the original notebooks. We also thank the AI4Life consortium for their support, and continous feedback.