A tool for visualizing Splotch results.
A custom version of Spav is used on https://als-st.nygenome.org.
Spav currently supports five different interactive views for visualizing and exploring Splotch results.
This view visualizes expression estimates on all the arrays simultaneously.
This view visualizes expression estimates on a selected array. The user can filter arrays based on the level 1 information (see Splotch).
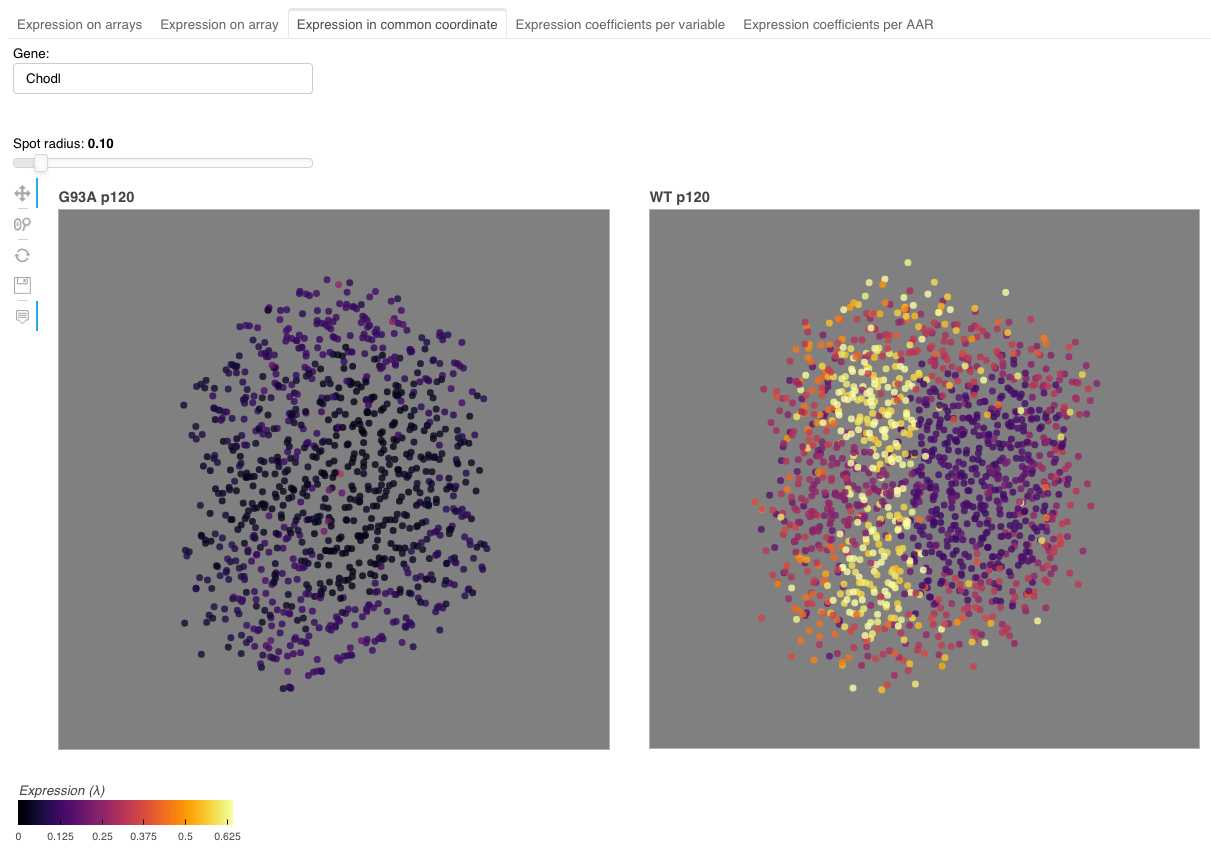
This view visualizes expression estimates in common coordinate system. The expression estimates are separated based on the level 1 information.
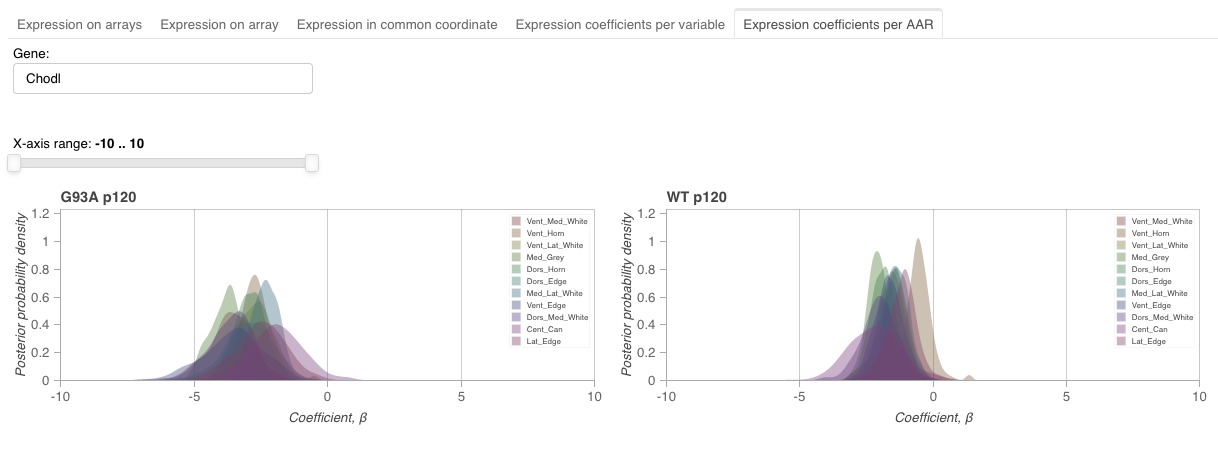
This view visualizes expression coefficients across level 1 variables for each AAR.
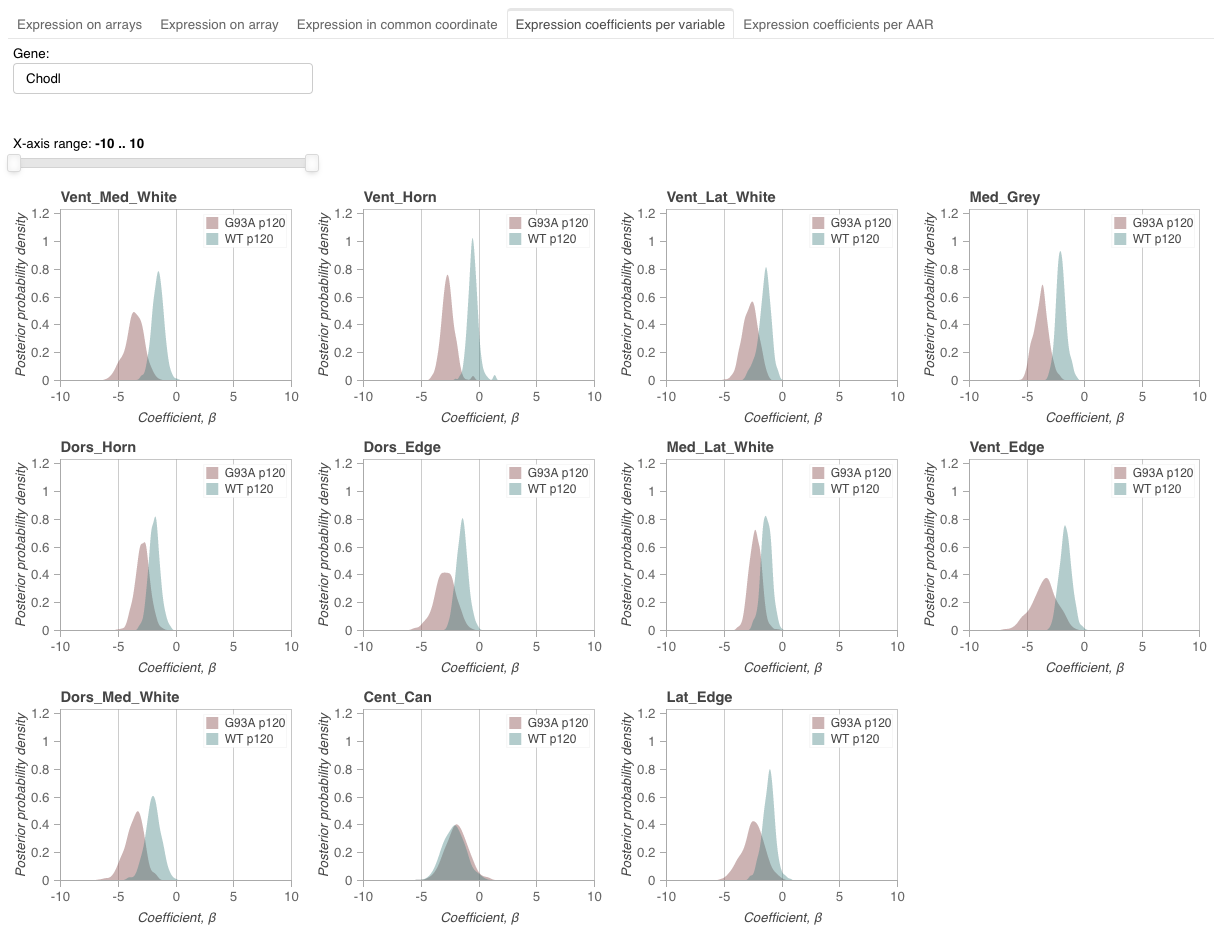
This view visualizes expression coefficients across AARs for each level 1 variable.
Spav has been tested on Python 3.7.
Spav can be installed as follows
$ pip install git+https://git@github.com/tare/Spav.gitThe script spav_prepare_data can be used for preparing the Splotch results to be used with Spav
$ spav_prepare_data --help
usage: spav_prepare_data [-h] -d DATA_DIRECTORY -o OUTPUT_DIRECTORY -s
SERVER_DIRECTORY [-c] [-v]
A script for preparing Splotch results for Span
optional arguments:
-h, --help show this help message and exit
-d DATA_DIRECTORY, --data-directory DATA_DIRECTORY
data directory
-o OUTPUT_DIRECTORY, --output-directory OUTPUT_DIRECTORY
output directory
-s SERVER_DIRECTORY, --server-directory SERVER_DIRECTORY
server directory
-c, --no-copy create symbolic links instead of copying images
-v, --version show program's version number and exitFor instance, if your Splotch input and output directories are $DATA_DIRECTORY and $OUTPUT_DIRECTORY (see Splotch), respectively, then you can prepare the data for Spav by executing the following command
$ spav_prepare_data -d $DATA_DIRECTORY -o $OUTPUT_DIRECTORY -s $SPAV_DIRECTORYThis command will create the directories $SPAV_DIRECTORY/static and $SPAV_DIRECTORY/data.
The directory $SPAV_DIRECTORY/static contains symbolic links pointing to the bright-field images and the $SPAV_DIRECTORY/data.hdf5 file contains the estimates.
For detailed description of how run a Bokeh server, please see https://bokeh.pydata.org/en/latest/docs/user_guide/server.html.
The script server/main.py implements our Bokeh application
$ python main.py --help
usage: main.py [-h] [--arrays] [--array] [--common-coordinate]
[--aar-coefficients] [--level-coefficients] [-v]
Spav server
optional arguments:
-h, --help show this help message and exit
--arrays show the array view
--array show the arrays view
--common-coordinate show the common coordinate view
--aar-coefficients show the aar coefficient view
--level-coefficients show the level coefficient view
-v, --version show program's version number and exitAdditionally, the directory server contains theme.yaml and templates/index.html.
First, let us copy the files from the directory server to the directory we created using the spav_prepare_data script
$ cp -r server/* $SPAV_DIRECTORY/ Second, the Bokeh application with all the implemented views can started by executing the following command
$ bokeh serve $SPAV_DIRECTORY --show --args --arrays --array --common-coordinate --aar-coefficients --level-coefficientsPlease see Notebook.ipynb.