Domain-interactive Contrastive Learning and Prototypeguided Self-training for Cross-domain Polyp Segmentation
Authors:
- This repository provides code for "Domain-interactive Contrastive Learning and Prototypeguided Self-training for Cross-domain Polyp Segmentation (DCLPS)" under review.
Accurate polyp segmentation plays a critical role from colonoscopy images in the diagnosis and treatment of colorectal cancer. While deep learning-based polyp segmentation models have made significant progress, they often suffer from performance degradation when applied to unseen target domain datasets collected from different imaging devices. To address this challenge, unsupervised domain adaptation (UDA) methods have gained attention by leveraging labeled source data and unlabeled target data to reduce the domain gap. However, existing UDA methods primarily focus on capturing class-wise representations, neglecting domain-wise representations. Additionally, uncertainty in pseudo labels could hinder the segmentation performance. To tackle these issues, we propose a novel Domain-interactive Contrastive Learning and Prototype-guided Self-training (DCL-PS) framework for cross-domain polyp segmentation. Specifically, domain-interactive contrastive learning (DCL) with a domain-mixed prototype updating strategy is proposed to discriminate class-wise feature representations across domains. Then, to enhance the feature extraction ability of the encoder, we present a contrastive learning-based cross-consistency training (CL-CCT) strategy, which is imposed on both the prototypes obtained by the outputs of the main decoder and perturbed auxiliary outputs. Furthermore, we propose a prototype-guided self-training (PS) strategy, which dynamically assigns a weight for each pixel during self-training, improving the quality of pseudo-labels and filtering out unreliable pixels. Experimental results demonstrate the superiority of DCL-PS in improving polyp segmentation performance in the target domain.
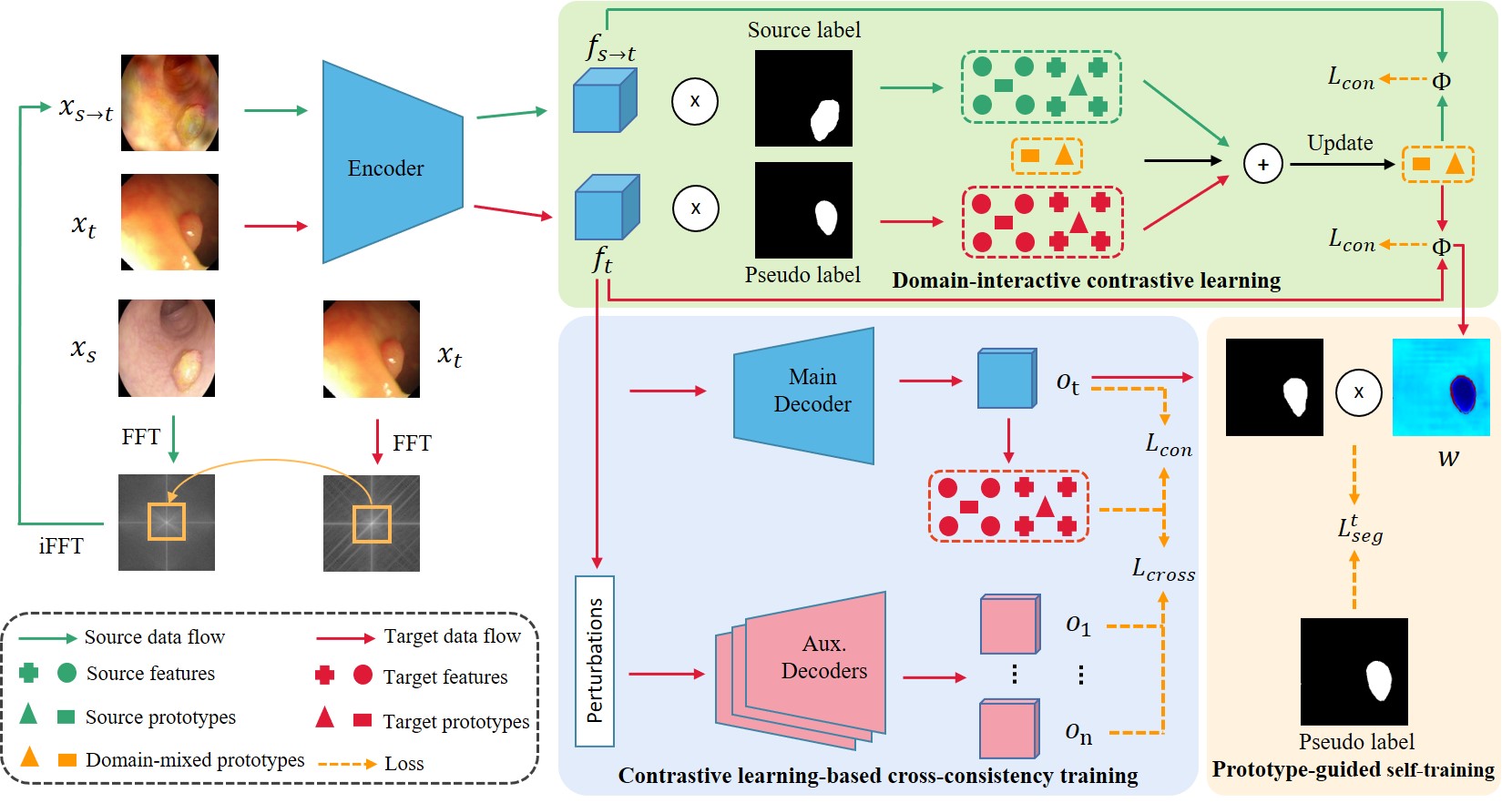
Figure 1: Overview of the proposed DCL-PS.
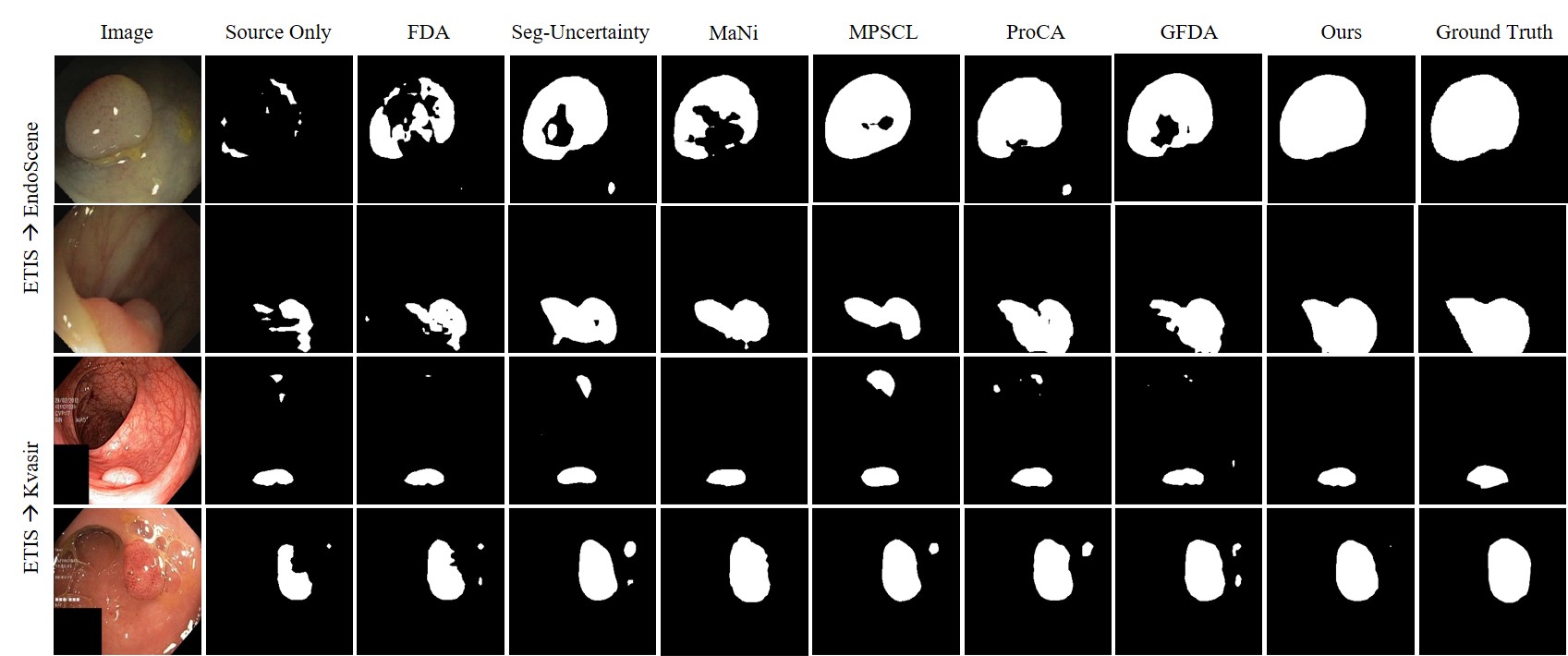
Figure 2: Qualitative Results.
The training and testing experiments are conducted using PyTorch with a single Nvidia GeForce 3090.
-
Configuring your environment (Prerequisites):
- Creating a virtual environment in terminal:
conda create -n DCLPS python=3.8 - Installing necessary packages:
pip install -r requirements.txt.
- Creating a virtual environment in terminal:
-
Downloading necessary data:
- DeepLab initialization can be downloaded through this line.
-
Training Configuration:
- just run:
sh run.shto train our model.
- just run:
-
Testing Configuration:
- Pre-computed maps will be uploaded later.
This code is heavily based on the open-source implementations from FDA and MPSCL
Please cite our paper if you find the work useful:
The source code is free for research and education use only. Any commercial use should get formal permission first.