This repository includes the codes and data for our HBeeID paper, titled "HBeeID: A molecular tool that identifies honey bee subspecies from different geographic populations," currently under review. Please find a list of included files as follows.
- SNPs_272_IDs_order.xlsx: A file that contains the list of the IDs for the 272 SNPs.
- Supp_Table_10_All_SNP_Genotype_Test_Datasets_for_HBeeID_10_3_22.xlsx: A table that contains SNP genotype data used as input to test HBeeID. Data from this work and from published datasets: Data for this work; Cridland et al. (2018); Avalos et al. (2017); Kadri et al. (2016); Wallberg et al. 2014; Harpur et al. (2014); Harpur et al. (2019). This table is also posted on the Github website associated with this publication. Harpur 2019 is missing from the table.
- Supp_Table_11_HBeeID_Assignations_Test_Datasets_with_Metadata_10_3_22.xlsx: A table that contains HBeeID assignations of samples from the data sets used to test HBeeID: Data from this work and published data sets: Cridland et al. (2018); Avalos et al. (2017); Kadri et al. (2016); Wallberg et al. 2014; Harpur et al. (2014); Harpur et al. (2019) along with their metadata. This table is also posted on the Github website associated with this publication.
- convertHaploidGenotypesInVCFFormat.R: A R script to convert haploid genotypes in VCF format to phased diplotized genotypes in VCF format.
- createDAPCfuncs_allNodes.R: A R Script to create DAPC model based on the SNP genotype data. This gives an example to create models based on user data by following the proposed workflow.
- dapc_funcs_AllNodes.RData: The DAPC model pre-generated by using the SNP genotype data in our paper.
- generateProportionalGraphs.R: A R Script to generate the proportional graphs in Figure 7 based on genotype data in Supplementary Table 4.
- predict_UN_pops.Rmd: A R Script to predict unknown samples by using generated DAPC model.
- Adegenet
- Readxl
- Openxlsx
- Tidyverse
- readr
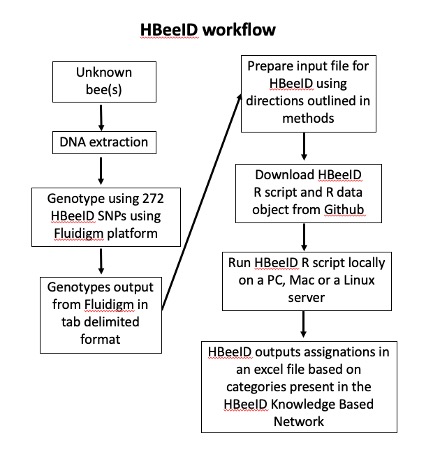
HBeeID requires as input genotype data in CSV format where the sample identification codes are represented in rows and the SNP genotypes are shown in columns. The first row should contain headers that describe the content of each column.
The first column should contain sample identification codes. There should be a total of 273 columns, where the first column is the sample identification codes column, followed by the SNP genotypes for the 272 SNPs in HBeeID.
The file, SNPs_272_IDs_order.xlsx, contains the list of the IDs for the 272 SNPs.
Each SNP ID contains the information of scaffold ID and the coordinate of the SNP loci in the Apis mellifera genome assembly, Amel4.5, available on BeeBase. The order of the SNPs in the input file must match the order shown in the file SNPs_272_IDs_order.xlsx.
The genotype data should be converted to [0, 1 or 2] where [0] represents the homozygote state for the reference allele, [1] represents the heterozygote state and [2] represents the homozygote state for the alternate allele.
The input file, prepared as specified above, needs to be given as input under the following line within the HBeeID code
unknowns_data <- read_csv("Input_genotypes.csv")
To facilitate easy testing of the HBeeID the following test datasets are provided with data extracted in the required input format of HBeeID as described above.
- Data from this work – 34 samples from PR
- Cridland et al. 2018 – 26 samples from Northern California and 18 samples from the Southern California
- Avalos et al. 2017 – 30 EHB samples from Hawaii; 28 AHB samples from Mexico; 30 PRHB samples from Puerto Rico
- Kadri et al. 2016 – 26 samples from Brazil
- Wallberg et al. 2014 – 10 A. m. adansonii samples, 10 Africanized samples from Brazil, 10 A. m. anatoliaca samples, 20 A. m. mellifera EU domestic, 10 A. m. mellifera US domestic, 10 A. m. carnica samples, 10 A. m. capensis samples, 10 A. m. Iberiensis samples, 10 A. m. ligustica samples, 20 A. m. mellifera Swedish-Norway samples, 10 A. m. scutellata samples
A subset of 183,609 SNPs in vcf format and gzipped. These are the SNPs common between Avalos et al (2017) and Wallberg et al (2014) SNP datasets and do not contain any missing genotypes in all 15 populations used in this study. These SNPs were analyzed with iterative DAPC analysis for the identification of the diagnostic SNPs.
The aforementioned SNP data are made available at Google Drive.
- Avalos A, Pan H, Li C, Acevedo-Gonzalez JP, Rendon G, Fields CJ, Brown PJ, Giray T, Robinson GE, Hudson ME, Zhang G. 2017. A soft selective sweep during rapid evolution of gentle behaviour in an Africanized honeybee. Nature Communications 8:1550.
- Cridland JM, Ramirez SR, Dean CA, Sciligo A, Tsutsui ND. 2018. Genome Sequencing of Museum Specimens Reveals Rapid Changes in the Genetic Composition of Honey Bees in California. Genome Biology and Evolution 10(2):458–472.
- Kadri SM, Harpur BA, Orsi RO, Zayed A. 2016. A variant reference data set for the Africanized honeybee. Scientific Data 3(1):160097
- Marcelino J, Donthu R, Avalos A, Weber R, Giordano R, Giray T. in prep. Impact of hurricane Maria on the genetic variation of honey bees in Puerto Rico.
- Wallberg A, Han F, Wellhagen G, Dahle B, Kawata M, Haddad N, Simões ZLP, Allsopp MH, Kandemir I, De la Rúa P, Pirk CW, Webster MT. 2014. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nature Genetics 46:1081–1088.
- Avalos et al. 2017 – NCBI BioProject PRJNA381313
- Cridland et al. 2018 – NCBI BioProject PRJNA385500
- Kadri et al. 2016 – NCBI BioProject PRJNA324081
- Wallberg et al. 2014 – NCBI BioProject PRJNA236426.