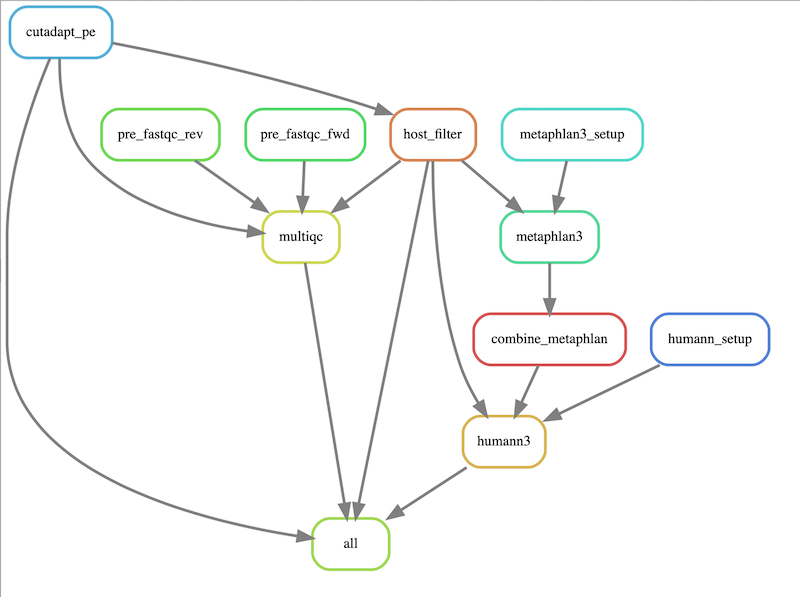
Stub workflow for a Snakemake metagenomics pipeline
We will be using this repository to develop a basic pipeline for metagenomic analysis using Snakemake.
The pipeline is meant to automate the processing and analysis of metagenomic data using HUMAnN3.
The workflow will execute the following steps:
- preliminary QC of reads using FastQC
- read trimming using Cutadapt
- human read removal using Bowtie2
- trimmed read QC using FastQC
- QC summary using MultiQC
- Taxonomic profiling using MetaPhlAn3
- Functional profiling using HUMAnN3
We will be using Snakemake to run our workflow for us. Snakemake is an absolutely amazing workflow language designed for scientific computing. It's especially suited to handling the kinds of data we generate with DNA sequencing -- many independent files, all of which need to undergo the same processing steps.
We'll follow the steps for installing Snakemake recommended on the website. The only difference is that we'll be installing Snakemake into a new environment dedicated for this metagenomic pipeline, so maybe we'll give it its own name. Here's what I did:
mamba create -c conda-forge -c bioconda -n quinn_mg snakemake
Now, we can activate this new environment and start programming!
(base) macbook:pkgs jgs286$ conda activate quinn_mg
(quinn_mg) macbook:pkgs jgs286$ snakemake --version
5.32.0
We will be using a tabular text file to tell Snakemake what are samples are, and any metadata associated with them about which we want it to know. In this case, the main thing is where the reads associated with the sample can be found!
Our samples.txt file will look something like this:
sample fq1 fq2
A test_data/S22205_S104_L001_R1_001.fastq.gz test_data/S22205_S104_L001_R2_001.fastq.gz
B test_data/S22207_S103_L001_R1_001.fastq.gz test_data/S22207_S103_L001_R2_001.fastq.gz