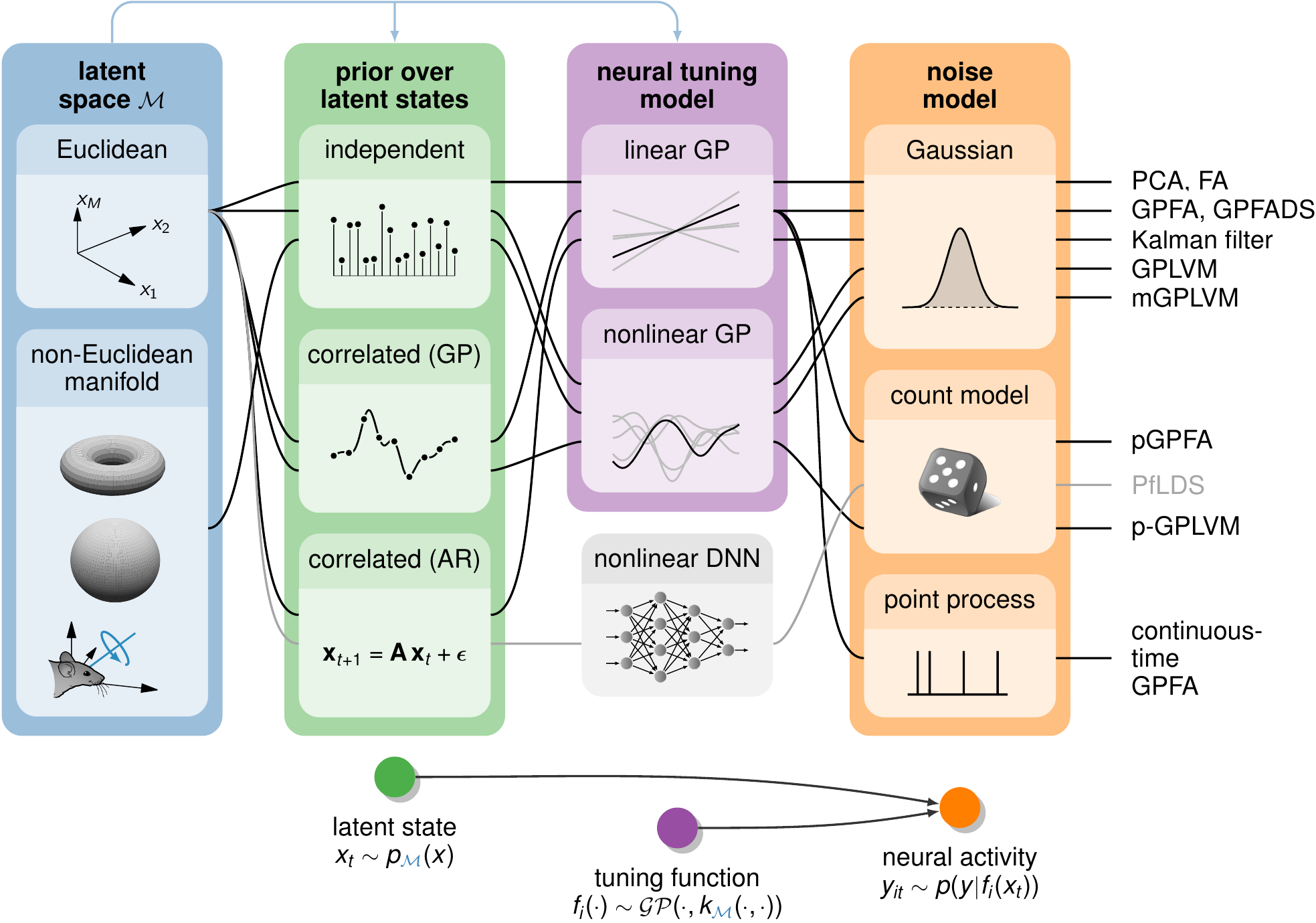
This repository contains code for running both the manifold GPLVM (Jensen et al. 2020) and Bayesian GPFA (Jensen and Kao et al. 2021) and is currently still in active development. In addition to these two recently published models, the codebase also faciliates the construction of a wide range of other models that combine different latent priors and linear/nonlinear tuning functions with Gaussian or discrete noise models (schematic) - all under the general framework of the Gaussian process latent variable model. The primary focus of this library is the analysis of neural population recordings, but the methods generalize to other domains as well.
To illustrate the use cases of these methods and provide a starting point for interested users, we have generated three example notebooks that run on Google Colab.
Bayesian GPFA with automatic relevance determination applied to recordings from monkey M1:
https://colab.research.google.com/drive/1Q-Qy8LM_Xn52g4dYycPRaBx0sMsti4_U?usp=sharing
mGPLVM for unsupervised learning on non-Euclidean manifolds, applied to data from the Drosophila head direction circuit:
https://colab.research.google.com/drive/1SoZGqYoPFSz-VQ6woo9QsoCzw3b1Pwut?usp=sharing
Adaption of mGPLVM for supervised learning on non-Euclidean manifolds, applied to synthetic data on the ring and group of 3D rotations:
https://colab.research.google.com/drive/1C7x5u4cMsH5f4i261Yz81zgDHgcJ-_MY?usp=sharing
# inside virtual environment
pip install -e .To run on GPU, it may be necessary to first install pytorch with GPU support.
- pytorch
- numpy
- scipy
- matplotlib
This library uses yapf for autoformatting. To autoformat all files in this directory:
yapf -ir .py.test@inproceedings{jensen2020manifold,
author = {Jensen, Kristopher and Kao, Ta-Chu and Tripodi, Marco and Hennequin, Guillaume},
booktitle = {Advances in Neural Information Processing Systems},
editor = {H. Larochelle and M. Ranzato and R. Hadsell and M. F. Balcan and H. Lin},
title = {Manifold {GPLVM}s for discovering non-{E}uclidean latent structure in neural data},
year = {2020},
}
@inproceedings{
jensen2021scalable,
title={Scalable {B}ayesian {GPFA} with automatic relevance determination and discrete noise models},
author={Kristopher T Jensen and Ta-Chu Kao and Jasmine Talia Stone and Guillaume Hennequin},
booktitle={Advances in Neural Information Processing Systems},
editor={A. Beygelzimer and Y. Dauphin and P. Liang and J. Wortman Vaughan},
year={2021},
}
@article{
jensen2022beyond,
title={Beyond the {E}uclidean brain: inferring non-{E}uclidean latent trajectories from spike trains},
author={Jensen, Kristopher T and Liu, David and Kao, Ta-Chu and Lengyel, M{\'a}t{\'e} and Hennequin, Guillaume},
journal={bioRxiv},
year={2022},
publisher={Cold Spring Harbor Laboratory}
}