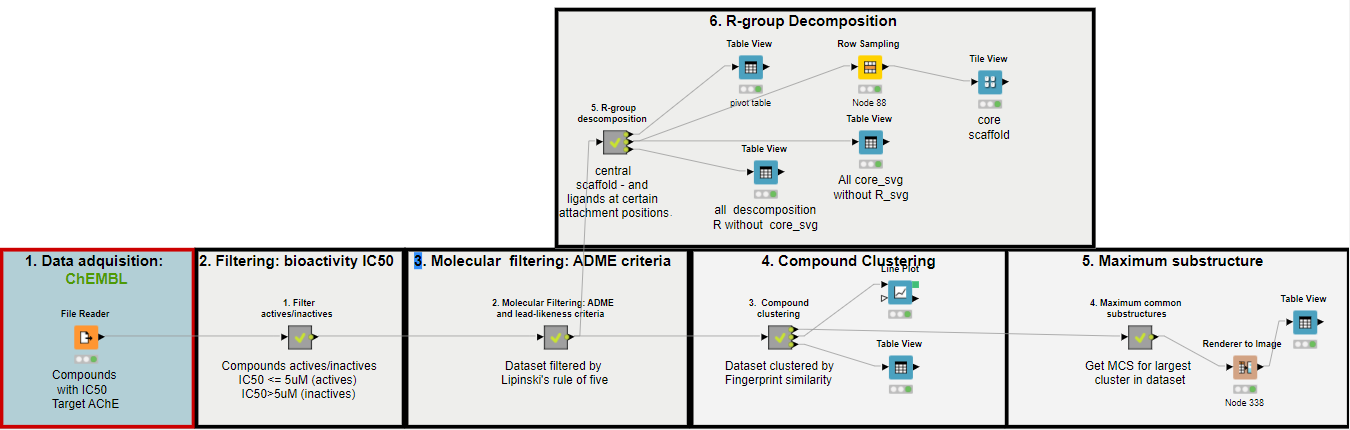
This workflow is composed of 5 interconnected blocks, each one on these containing a central theme in computer-aided drug design (CADD).
The workflow is presented using Acetylcholinesterase (AChE) as an example, including its respective active compounds and their respective associated IC50 values, which where obtained from the CHEMBL database. Our chemoinformatics analysis allows the user to:
- List active and inactive compounds (according to the selected IC50 threshold).
- Evaluate the compounds bioavailability and other properties which adversely affect the absorption, distribution, metabolism, and excretion (ADME) of those compounds, based on chemical structure alone.
- Group compounds according to their structural similarity in order to expose activity patterns, or for novel compound construction through scaffold fusion. From this grouping, a diverse set of compounds can also be selected for further analysis.
- Calculate the common maximum substructure in a set of molecules.
- Find a central structure (scaffold) within all active compounds, and identify its substituents at certain binding positions.
- Knime version 4.3.1, an open source software for data analysis Knime website.
- Our Knime workflow, to capture relevant data about an indication of interest making use of different databases Chemoinformatics-analysis.
- Input files Inputs.
- Here you can have a complete visualization of the workflow Workflow Visualization.
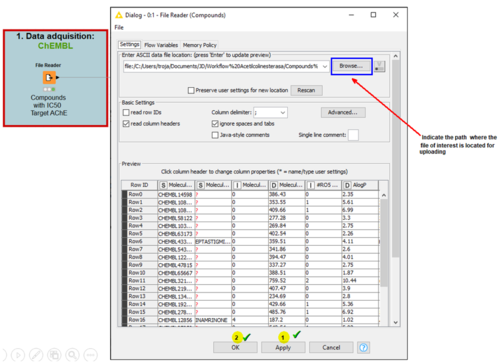
1. Data acquisition from CHEMBL
Information related to the structure, bioactivity, and targets associated to a compound are available in databases such as CHEMBL, PubChem, or DrugBank. This data is obtained from CHEMBL where all compounds associated with the chosen target (Acetylcholinesterase AChE) which had an IC50 value, were filtered along with their respective Smiles and phase of study.
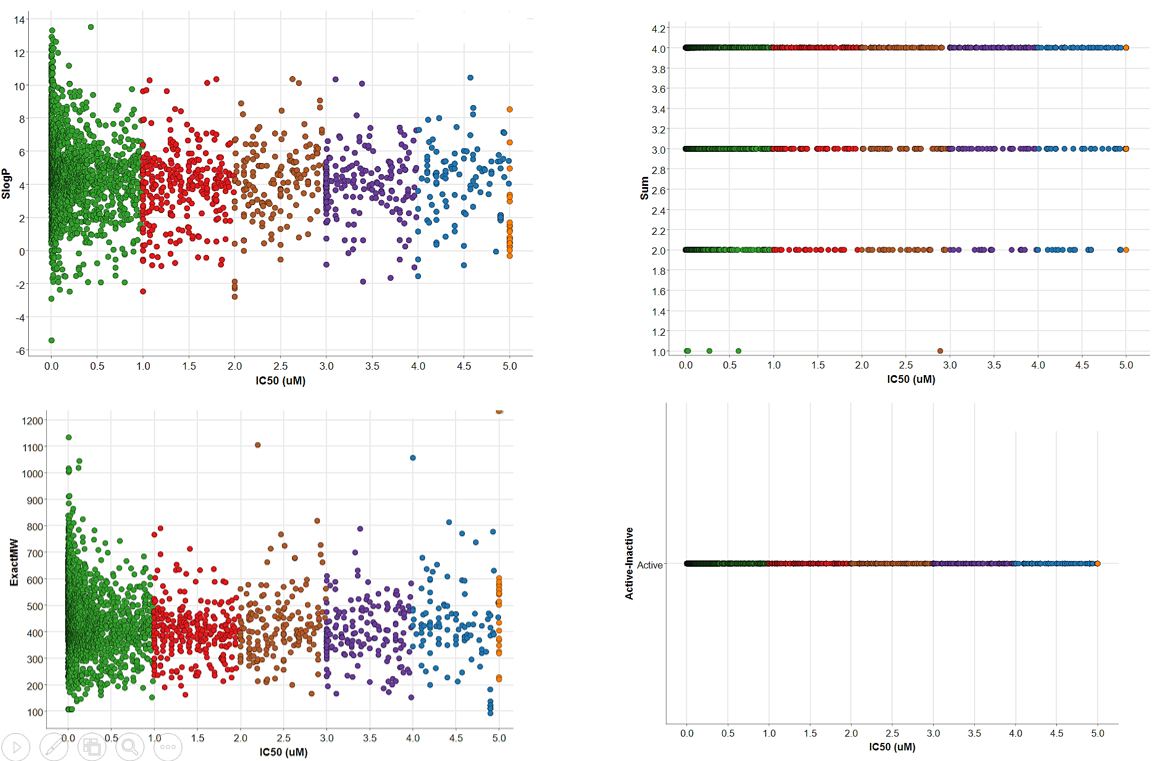
Active compounds were separated from inactive compounds through the following parameter:
Active IC50 < 5 µM
Inactive IC50 > 5 µM
Not all compounds are suitable as a starting point for drug development due to their undesirable pharmacokinetic properties, which could adversely affect the absorption, distribution, metabolism, and excretion (ADME) of a drug. Therefore, these compounds are usually excluded in datasets aiming to target promising drug candidates. Therefore, fewer drug-like molecules in the dataset should be eliminated.
The bioavailability of a compound is an important ADME property. Lipinski's rule of five (RO5) was introduced to estimate the bioavailability of a compound based solely on their chemical structure. RO5 states that malabsorption or permeation of a compound is more likely to occur if the chemical structure violates more than one of the following rules:
- Molecular weight (MWT) <= less than 500 Da.
- Number of hydrogen bond acceptors (HBA) <= no more than 10.
- Number of hydrogen bond donors (HBD) <= 5 or less.
- Calculated LogP (octanol-water coefficient) <= 5 or less.
- Subsequently, those compounds complying with three or four of Lipinski's rules were filtered out before further chemometric analyses.
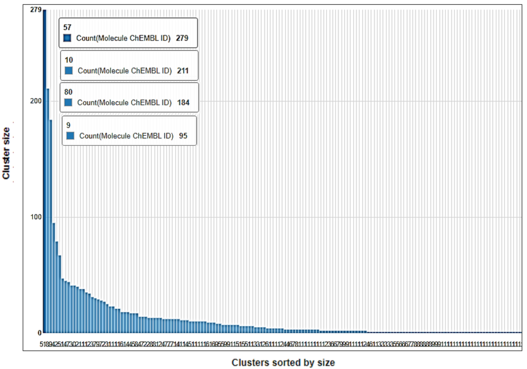
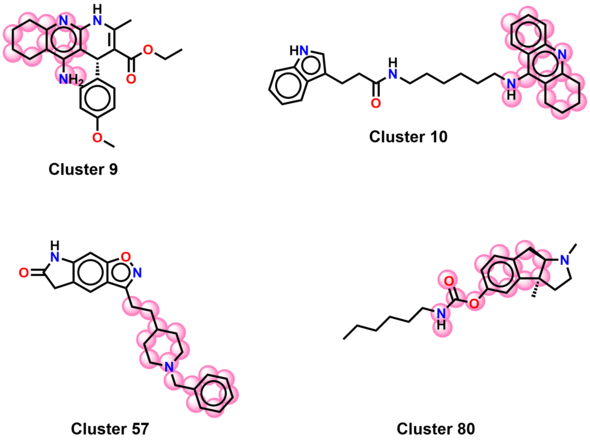
Clustering can be used to identify groups composed of similar compounds, in order to pick a set of diverse compounds from these clusters for diverse goals like, for exmaple, non-redundant experimental testing. The following steps show how to create these clusters based on a hierarchical clustering algorithm.
To visualize the shared scaffolds between compounds, and thus emphasize the extent and type of chemical similarities inside a cluster, the maximum common substructure (MCS) can be calculated and highlighted. In this workflow, the MCS was calculated for the four significant clusters obtained from the previous node using the FMCS algorithm.
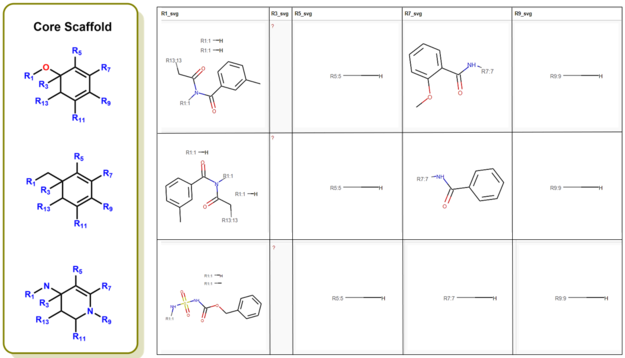
R-group decomposition is a special type of search that aims to find a central substructure (scaffold) and identify its substituents at certain binding positions. The query molecule consists of the scaffold and binding sites represented by R-groups.
This block of the workflow shows how to perform R-group decomposition using the RDKit community extension. Its implementation consists of several steps.
- Calculate MCS taking all active compounds as a starting point.
- Perform the R-group decomposition.
- Find how many molecules with each combination of the two selected R-groups are in the dataset.
- Visualize the results of the R-group decomposition.
- Hurtado-Pachon, Jonathan; Peña-Varas, Carlos; Ramírez, David. Chemoinformatics analysis for computer-aided drug design (CADD). Santiago, Chile; 2021.