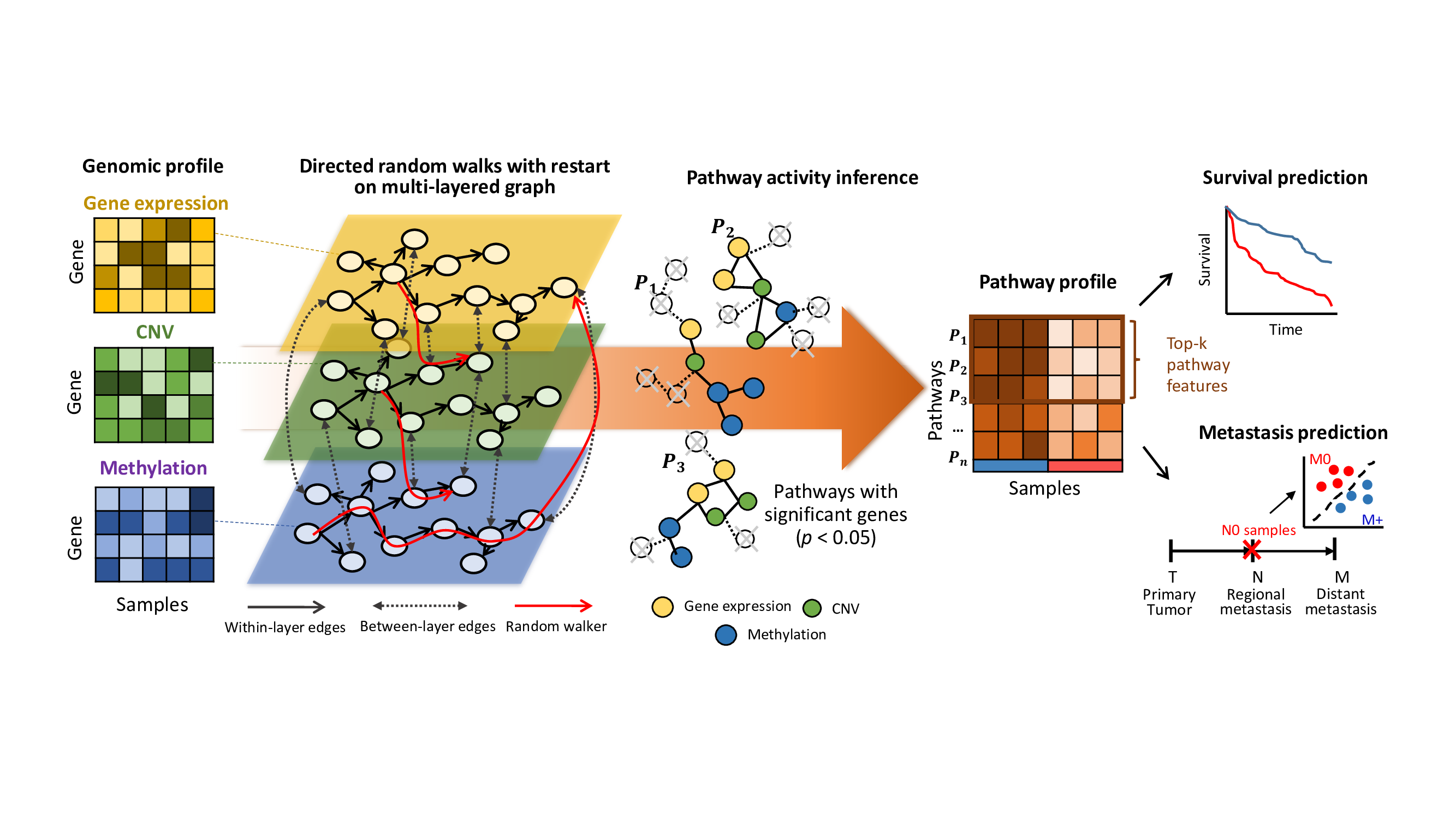
iDRW is an integrative pathway activity inference method using directed random walks on graph. It integrates multiple genomic profiles and transfroms them into a single pathway profile using a pathway-based integrated gene-gene graph.
library(devtools)
install_github("sykim122/iDRW")library(iDRW)Try our sample data (TCGA Bladder cancer dataset) and KEGG pathway-based gene-gene graph
data("data_BLCA")data_BLCA contains have three genomic profiles and clinical matrix.
exp: RNA-Seq gene expression profilecna: CNV profilemeth: DNA methylation profileclinical: clinical matrix (7 variables -time(overall survival days),status(event status),age,gender,stageM,stageN,stageT)
directGraph and pathSet contain directed gene-gene graph (igraph object) and the list of KEGG pathways.
Now, construct three-layered gene-gene graph from sample data.
library(igraph)
data("directGraph.KEGGgraph")
data("pathSet.KEGGgraph")
g <- directGraph
c <- directGraph
m <- directGraphGenes should be named with delimiters as below.
gene_delim <- c('g.', 'c.', 'm.') # genes from RNA-Seq gene expression(g), CNV(c), Methylation(m) profile
V(g)$name <- paste(gene_delim[1],V(g)$name,sep="")
V(c)$name <-paste(gene_delim[2],V(c)$name,sep="")
V(m)$name <-paste(gene_delim[3],V(m)$name,sep="")Initially, multi-layered graph simply can be constructed by the union of three graphs (the within-layer interactions are defined in directGraph). The between-layer interactions will be assigned in Step 3.
gcm <- (g %du% c) %du% mIn this example, we select significant genes associated with survival outcome by a univariate cox regression model, adjusted by age, gender, TNM stage.
class.outcome <- "time"
covs <- c("age", "gender", "stageT", "stageN", "stageM")
family <- "cox"
pa <- get.iDRWP(x=list(exp, cna, methyl), y=clinical, globalGraph=gcm, pathSet=pathSet, class.outcome=class.outcome,
covs=covs, family=family, Gamma=0.3, Corr=FALSE) pa$pathActivity is a pathway profile inferred by iDRW (samples x pathways).
For more information, please refer the following document with ?get.iDRWP or help(get.iDRWP).
Please cite our papers if you use this package in your own work.
@article{kim2020multi,
title={Multi-layered network-based pathway activity inference using directed random walks: application to predicting clinical outcomes in urologic cancer},
author={Kim, So Yeon and Choe, Eun Kyung and Shivakumar, Manu and Kim, Dokyoon and Sohn, Kyung-Ah},
journal={bioRxiv},
year={2020}
}
@article{kim2019robust,
title={Robust pathway-based multi-omics data integration using directed random walks for survival prediction in multiple cancer studies},
author={Kim, So Yeon and Jeong, Hyun-Hwan and Kim, Jaesik and Moon, Jeong-Hyeon and Sohn, Kyung-Ah},
journal={Biology direct},
volume={14},
number={1},
pages={1--13},
year={2019},
publisher={BioMed Central}
}
@article{kim2018integrative,
title={Integrative pathway-based survival prediction utilizing the interaction between gene expression and DNA methylation in breast cancer},
author={Kim, So Yeon and Kim, Tae Rim and Jeong, Hyun-Hwan and Sohn, Kyung-Ah},
journal={BMC medical genomics},
volume={11},
number={3},
pages={33--43},
year={2018},
publisher={BioMed Central}
}
So Yeon Kim jebi1771@gmail.com