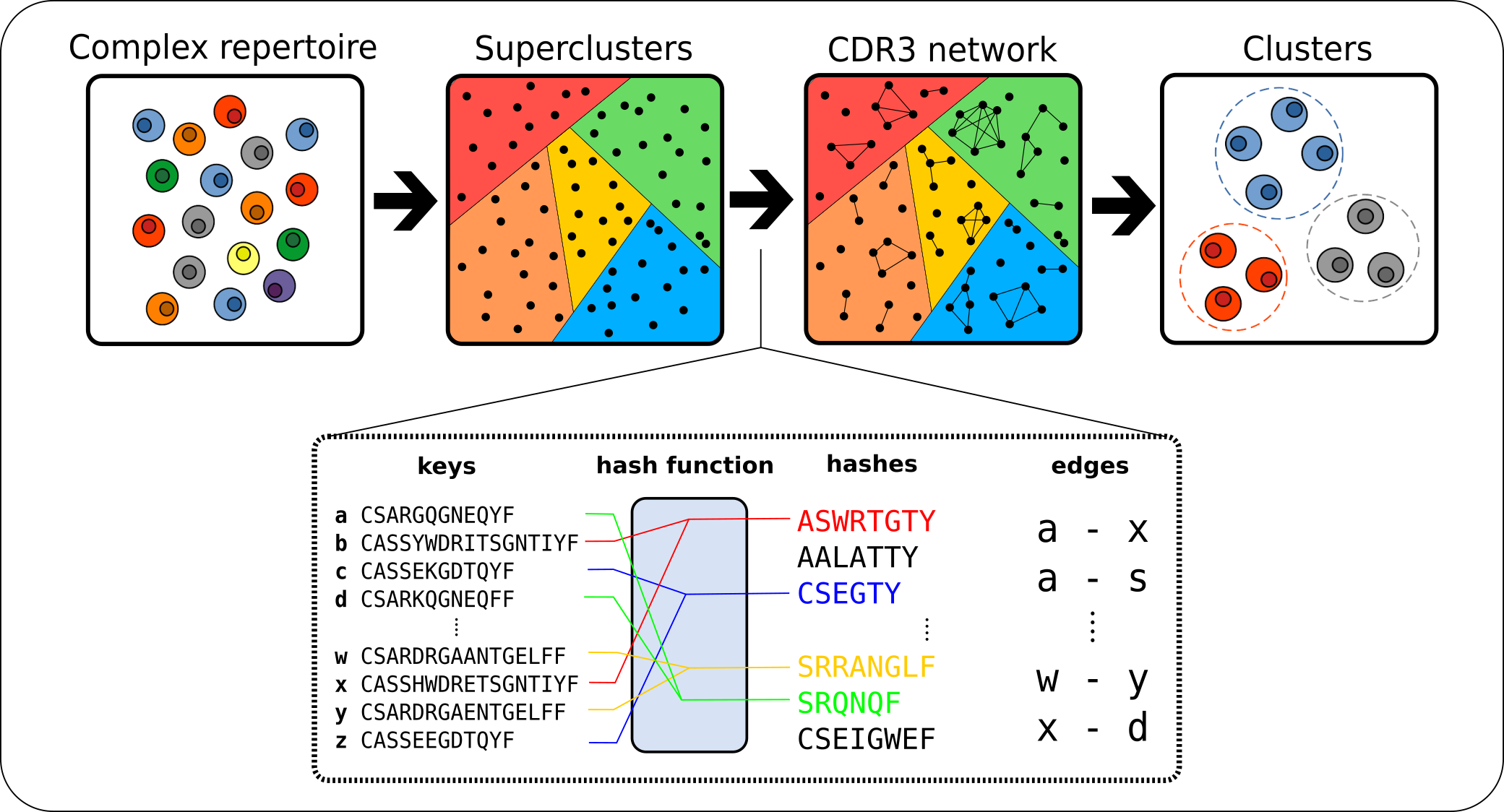
ClusTCR: a Python interface for rapid clustering of large sets of CDR3 sequences with unknown antigen specificity 

A two-step clustering approach that combines the speed of the Faiss Clustering Library with the accuracy of Markov Clustering Algorithm
On a standard machine*, clusTCR can cluster 1 million CDR3 sequences in under 5 minutes.
*Intel(R) Core(TM) i7-10875H CPU @ 2.30GHz, using 8 CPUs
Compared to other state-of-the-art clustering algorithms (GLIPH2, iSMART and tcrdist), clusTCR shows comparable clustering quality, but provides a steep increase in speed and scalability.
Documentation & Install
All of our documentation, installation info and examples can be found in the above link! To get you started, here's how to install clusTCR
$ conda install clustcr -c svalkiers -c bioconda -c pytorch -c conda-forge
There's also a GPU version available, with support for the use_gpu parameter in the Clustering interface.
$ conda install clustcr-gpu -c svalkiers -c bioconda -c pytorch -c conda-forge
Mind that this is for specific GPUs only, see our docs for more information.
To update use a similar command
$ conda update clustcr -c svalkiers -c bioconda -c pytorch -c conda-forge
To start developing, after cloning the repository, create the necessary environment
$ conda env create -f conda/env.yml
The requirements are slightly different for the GPU supported version
$ conda env create -f conda/env_gpu.yml
To build a new conda package, conda build is used.
Mind that the correct channels (pytorch, bioconda & conda-forge) should be added first or be
incorporated in the commands as can be seen in the install commands above.
$ conda build conda/clustcr/
For the GPU package:
$ conda build conda/clustcr-gpu/
Please cite as:
Sebastiaan Valkiers, Max Van Houcke, Kris Laukens, Pieter Meysman, ClusTCR: a Python interface for rapid clustering of large sets of CDR3 sequences with unknown antigen specificity, Bioinformatics, 2021;, btab446, https://doi.org/10.1093/bioinformatics/btab446
Bibtex:
@article{valkiers2021clustcr,
author = {Valkiers, Sebastiaan and Van Houcke, Max and Laukens, Kris and Meysman, Pieter},
title = "{ClusTCR: a Python interface for rapid clustering of large sets of CDR3 sequences with unknown antigen specificity}",
journal = {Bioinformatics},
year = {2021},
month = {06},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btab446},
url = {https://doi.org/10.1093/bioinformatics/btab446},
note = {btab446},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btab446/38660282/btab446.pdf},
}