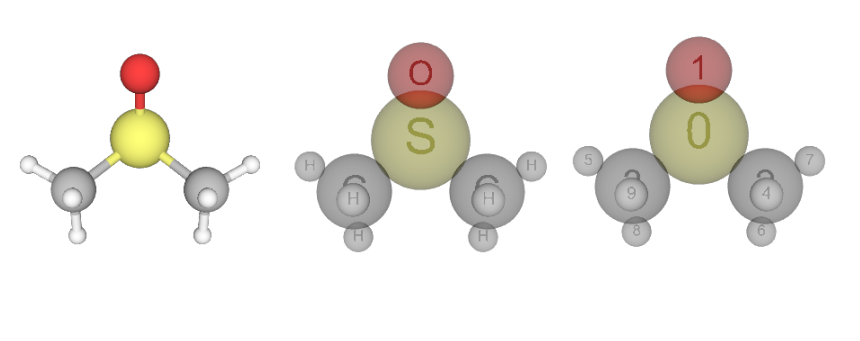
Python module for drawing and rendering atoms and molecules objects using X3DOM. X3dase can be used as a viewer for the molecule structure in the Jupyter notebook.
Functions:
- Support all file-formats using by ASE, including cif, xyz, cube, pdb, json, VASP-out and so on.
- Ball & stick
- Space filling
- Polyhedral
- Isosurface
- Show element and index
- Measure distance and angle
- Animation
For the introduction of ASE , please visit https://wiki.fysik.dtu.dk/ase/index.html
- Xing Wang xingwang1991@gmail.com
- Python
- ASE
- Skimage
pip install --upgrade --user x3daseYou can get the source using git:
git clone https://github.com/superstar54/x3dase.gitThen add it to your PYTHONPATH and PATH. On windows, you can edit the system environment variables.
export PYTHONPATH=/path-to-x3dase:$PYTHONPATH| key | function |
|---|---|
| b | ball-and-stick model |
| s | spacefilling model |
| p | polyhedra model |
| 1 | view top |
| 2 | view front |
| 3 | view right |
| 4 | view element |
| 5 | view index |
Using Ctrl + click to select atoms.
| Selection | measurement |
|---|---|
| single atom | xyz position and atomic symbol |
| two atoms | interatomic distance |
| three atoms | three internal angles |
images = [atoms1, atoms2, atoms3]
X3D(images)