- Name: Tutorial on adding Signposting to HTML in Web pages
- Description: This tutorial shows how to add Signposting to GitHub pages. It uses a simple GitHub page hosted in the
docs/folder to create a sample project page, i.e., as learners could do with their own GitHub projects. As an example, it uses the dataset corresponding to the released project TREC-doc-2-doc-relevance, a web-based interface to add document-to-document relevance assessments to pairs of documents retrieved from TREC 2005 Genomics Track. - Keywords: Signposting, GitHub pages
- How can I add Signposting to GitHub pages?
- How can I include external metadata in my signposting?
- How do I decide which metadata to include in signposting?
- Describe how Signposting can be embedded in GitHub pages
- Understanding of Signposting limitation of static content-delivery networks
- Knowledge of different metadata formats and their signposting profiles
- Familiarity on how to use GitHub
- Basic knowledge on how to use GitHub Pages
- Brief understanding of Signposting (introduction slides)
- Familiarity with HTML
- Knowledge of developer tools on a browser
Latest modification 2024-02-25
License CC-By 4.0
In this tutorial we will cover:
- signposting-tutorial
- Overview
- Learning experience
- Agenda
- Prerequisites
- Creating this GitHub Page
- Overview of the repository
- Challenge of machine actionability
- Adding FAIR Signposting
- Where to add Signposting?
- Adding a persistent identifier
- Specifying the resource type
- Specifying authors
- Specifying license
- Specifying content downloads
- Listing metadata resources
- Linking back to the collection
- Repository listing
- Linksets
- Try it out
- Acknowledgements
To follow this tutorial, you should already have experience with using GitHub, and be signed in to your GitHub account. See Learn the basics of GitHub as a refresher.
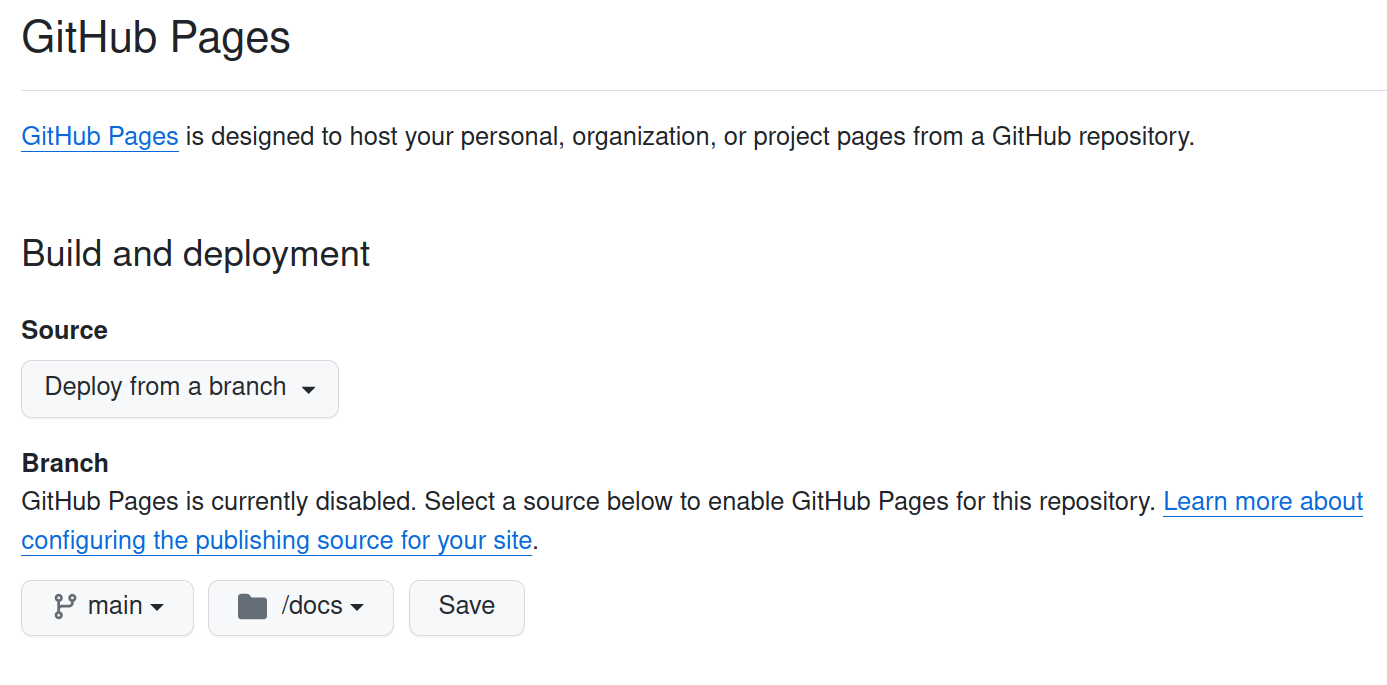
Let's start by forking this repository for your own purposes. Once forked, go to Settings
You will need to enable Pages on your forked repository, and select Deploy from a branch using Branch:
main and Folder: docs/. Then Save the changes.
As this repo does have a gh-pages branch, it will use it. If such branch would not exist, GitHub would ask you to use the main branch to start the gh-pages one
In a matter of minutes, your site will be live. The pages corresponding to the examples used in this tutorial are available at https://stain.github.io/signposting-tutorial/ and corresponding pages should appear by replacing stain with your GitHub username.
Do not forget to check out a local copy of your fork so you can make changes -- alternatively you may use the GitHub editor.
This repository is emulating a basic HTML-based institutional repository, with a single dataset entry corresponding to a Zenodo's entry. Layout:
- docs/ The Web root, as deployed above. Note that your deployment will use a hostname similar to
stain.github.iobut reflecting your username.- docs/7338056/ A single dataset, based on a real Zenodo entry
- docs/7338056/index.html HTML for the dataset 7338056, at start of tutorial without any signposting
- docs/7338056/solution.html Modified
index.htmlafter following this tutorial. Do not peek here until you have done the exercises! - docs/7338056/bioschemas.jsonld Bioschemas JSON-LD metadata as in the tutorial bioschemas-ghpages-markup-tutorial, but extracted from
<script>tag - docs/7338056/fleiss.tsv a file in tabuler-separated value format (converted from CSV)
- docs/7338056/ A single dataset, based on a real Zenodo entry
The remaining dataset and metadata downloads are in this case shown as deeplinks to Zenodo to indicate that Signposting is not tied to a particular domain.
This tutorial is deployed using GitHub Pages as described above. For simplicity it uses static HTML files based on a Bootstrap v5 starter template -- applying Signposting to a real repository deployment may require editing its HTML templates, which is currently out of scope for this tutorial.
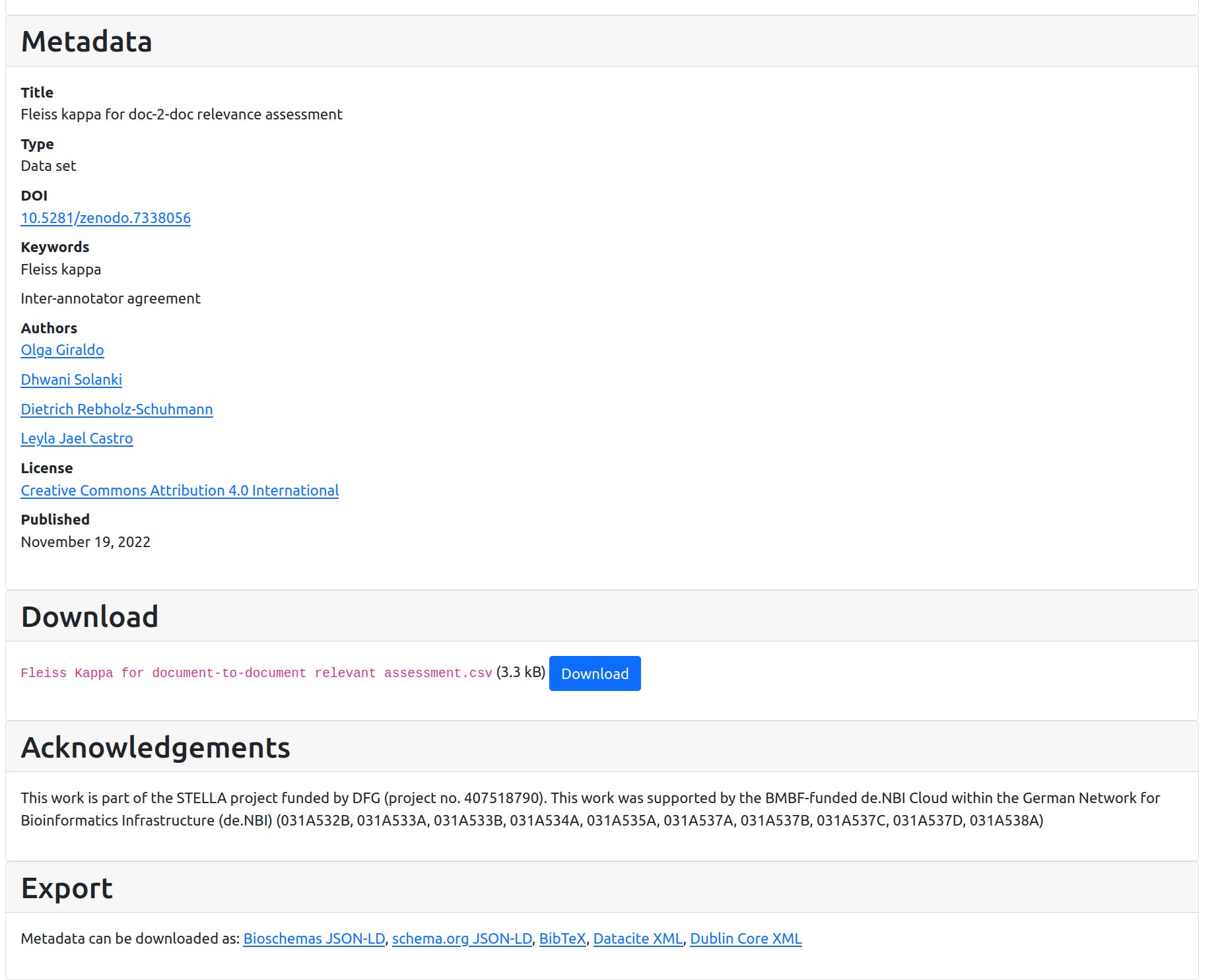
Look at HTML page https://stain.github.io/signposting-tutorial/7338056/ (or equivalent for your username) and open the HTML code in docs/7338056/index.html. This is a somewhat typical landing page for a Web-based data repository. We will imagine that the persistent identifier (DOI) has redirected to this page, as is the case for the original https://doi.org/10.5281/zenodo.7338056
We see that the landing page is quite useful for humans, including an abstract; metadata including title, author, keywords; and a big download button. There are some export formats listed at the end for metadata formats like Bibtex.
The tutorial bioschemas-ghpages-markup-tutorial highlights how this kind of metadata can be made machine-readable in a FAIR format -- which for completeness is included in the <script> tag at the end of the HTML. Bioschemas is however just one of the many ways that FAIR metadata can be provided (as shown in this example).
However, a machine (example: pre-programmed script) who accesses the given persistent identifier, and do not already know this particular repository implementation or Bioschemas, is not immediately able to answer the most basic FAIR questions:
- What is the persistent identifier?
- What is the type of the resource described?
- Where can it download the data (if any), and in which format(s)?
- What is the license and authorship of the data?
- What other metadata formats are available? What conventions do they follow?
The goal of Signposting is to reduce the heuristics that would otherwise be needed by such clients (e.g. text mining or content-negotiation), to give explicit typed links to facilitate navigation. Note that this is different from semantics, as the main goal is to give the client further waypoints rather than meaning.
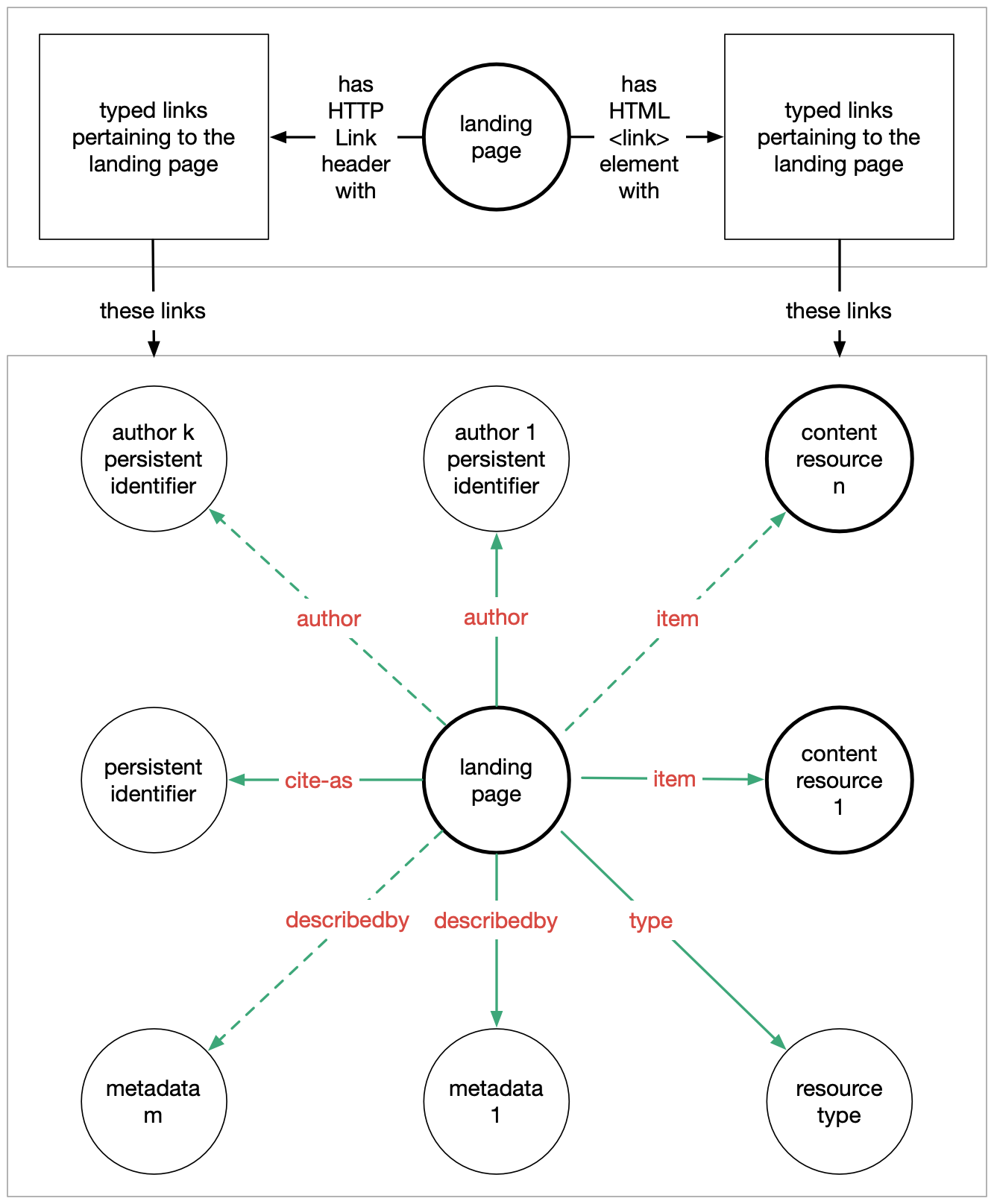
In this tutorial we will implement FAIR Signposting at level 1, which provides:
- Author(s)
- Persistent identifiers
- Metadata
- Download/archive
- License
- Type
Signposting can be added in three ways:
- In the HTTP
GET/HEADresponse, usingLinkheader - In a HTML landing page, within the HTML
<head>section using<link>element - In a dedicated linkset JSON or text file, linked to using any of the above
As this tutorial is neutral to deployment, and GitHub Pages do not permit control over HTTP headers, we will primarily work with option 2.
To add the HTML links to your forked repository, now open docs/7338056/index.html and click either Edit in place button or the more powerful Open with github.dev.
If you don't see these options, make sure you are on your fork of the repository.
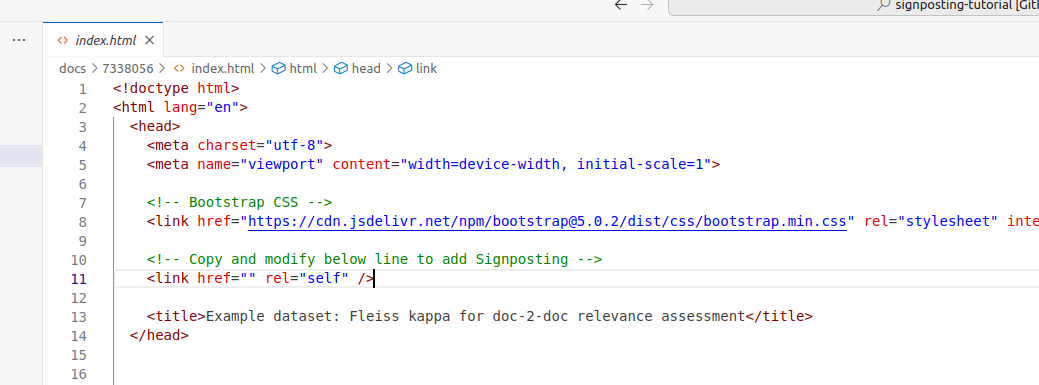
Towards the top of the file, you will find two tags we will expand:
<!-- Bootstrap CSS -->
<link href="https://cdn.jsdelivr.net/npm/bootstrap@5.0.2/dist/css/bootstrap.min.css"
rel="stylesheet" integrity="sha384-EVSTQN3/..." crossorigin="anonymous">
<!-- Copy and modify below line to add Signposting -->
<link href="" rel="self" />The first line shows how we are using the existing HTML mechanism for linking, rel=stylesheet tells the browser how to add the styling using the linked Bootstrap theme.
The second line is a template which we'll copy and modify in the instructions below. In the end you may delete this example line, as rel=self is not needed in HTML documents.
Make sure you add the new links within the <head> .... </head> section, as recommended by FAIR Signposting. To simplify life for clients, it is NOT RECOMMENDED to add <link> to the <body> content.
The FAIR Guiding Principles include:
F1. (meta)data are assigned a globally unique and persistent identifier
...
F3. metadata clearly and explicitly include the identifier of the data it describes
Persistent identifiers as expressed in Signposting using rel="cite-as" (RFC8574) -- this allows a landing page to say which persistent identifier will redirect to the page.
The original entry for this dataset has a DOI 10.5281/zenodo.7338056 -- however DOIs as untyped strings are not a good targets, as every Signposting has to be a valid URI -- typically starting with http:// or https:// followed by a domain name for the corresponding Web server. For DOIs we will therefore use the https://doi.org/ resolver -- to convert the DOI to a URI, simply add this as a prefix to become: https://doi.org/10.5281/zenodo.7338056
Modify docs/7338056/index.html so that it includes the signposting for the DOI
10.5281/zenodo.7338056:
<link href="https://doi.org/10.5281/zenodo.7338056" rel="cite-as" />Note however that the purpose of cite-as is not to give any odd scholarly citation, but a persistent identifier that leads back to this place. In this idealized example we have duplicated a Zenodo entry, however their DOI https://doi.org/10.5281/zenodo.7338056 of course will still redirect to their landing page https://zenodo.org/records/7338056 and we are not at power to modify their HTML template.
alt text
https://w3id.org/signposting-tutorial/{user}.{number}
Add the below signposting to reflect your username, and use this instead as a cite-as:
<link href="https://w3id.org/signposting-tutorial/stain.7338056" rel="cite-as" /> If you manage a repository, you likely already assign persistent identifiers that can be used with
cite-as -- if not, consider these resources:
- Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data
- DataCite: Create DOIs
- w3id: Permanent identifiers for the Web
- b2handle
The FAIR Signposting requires a type to classify the scholarly object, in our case a CSV file.
Browse the Schema.org hierarchy to expand
CreativeWork and find the type Dataset (other common types may be ScholarlyArticle, ImageObject, SoftwareSourceCode)
To specify
Dataset as a type, use:
<link href="https://schema.org/Dataset" rel="type" />Note: This schema.org identifier is subtly different from the JSON-LD usage in Bioschemas, which @context maps Dataset to http://schema.org/Dataset etc. As Signposting is navigational and not semantic, we here prefer the https:// variant.
Now, the resource we are providing the signposting from is not technically speaking the dataset, but a landing page about the downloadable dataset. Therefore Signposting recommends also adding:
<link href="https://schema.org/AboutPage" rel="type" /> This may be a good time to try it out using a signposting client to verify your changes to index.html.
If each author of the resource have some persistent identifier (e.g. ORCID), or other user page within the repository, we can list them using author link relation.
Add for each of the authors listed in the HTML their ORCID identifier using
rel="author":
<link href="https://orcid.org/0000-0003-2978-8922" rel="author" />Note that if the author does not have a page but only a name, you can't provide a link nor persistent identifier, and so there is nothing to signpost to. Remember the purpose here is navigation, full semantics is however left in the metadata, which we'll cover later.
In many cases, a repository entry has an open access or open source license. In this case it is very valuable to provide the license signposting, in order to indicate to clients what they are permitted to do with the download.
In our first attempt, let's specify the Creative Commons CC-BY 4.0 license by using the URI as provided in the HTML:
<link href="https://creativecommons.org/licenses/by/4.0/" rel="license" />Needless to say, there are many possible license, each of which may have many identifiers. So while this link may be useful for humans, for machine actionability it is preferrably to use a known persistent identifier also for the license.
The SPDX License List is such a well known set of license identifiers. Identify the line for "Creative Commons Attribution 4.0 International". Remember signposting can't go to untyped identifiers like
CC-BY-4.0 but needs a URI. Luckily SPDX provides such URIs e.g. https://spdx.org/licenses/CC-BY-4.0 (although, for unexplained reasons, their list links to .html variants).
Modify the above
license to use the SPDX persistent identifier:
<link href="https://spdx.org/licenses/CC-BY-4.0" rel="license" />In other cases there is no single license, or the license is only embedded within the dataset. In this case you should not include a license as you don't have a single resource to link to.
Tip: Make sure you use the US spelling of the link relation license!
Returning to the FAIR Principles we also find:
A1. (meta)data are retrievable by their identifier using a standardized communications protocol
If we accept that many persistent identifier goes to a HTML landing page, rather than directly to the downloadable data (which would then hide the metadata), A1 must be enabled for machine through an indirection. In Signposting this is done using the item link relation.
From the existing HTML we find the CSV file as a Download link.
Add the signposting for the download:
<link
href="https://zenodo.org/records/7338056/files/Fleiss%20Kappa%20for%20document-to-document%20relevant%20assessment.csv?download=1"
rel="item"
type="text/csv" />Note that although type is optional, it is strongly recommended for downloads, specially if the server is unable to return a correct Content-Type.
See the IANA media types or PRONOM to find known file formats.
It is possible to have additional downloads. For instance, Zenodo entries can have multiple uploads for a single DOI/landing page. In this tutorial repository, we have included the fleiss.tsv as an example of an additional resource, converted from the CSV to the Tabular Separated Values format .
Add another download for our converted TSV file:
<link
href="fleiss.tsv"
rel="item"
type="text/tab-separated-values" />Note: There is no indication in the outgoing links that these are alternatives of the same resource (the underlying table). This could have to be done using rel=alternate at a HTTP header level for each of the files, however this kind of semantics is not required by Signposting. Likewise, provenance history of a conversion taking place would be the role of metadata to cover.
A very important motivation for FAIR Signposting is to make machine-readable metadata easier to find. In particular, clients should not need to content-negotiate or know in advance exactly which format are available. In some cases metadata is also available externally, which is examplified by this repository, which links back to Zenodo.
In Signposting, metadata resources are listed as describedby. Metadata is considered separated from the data (the item downloads) if they can be considered to primarily be describing the data.
Note: A dataset may just happen to be written in a semantic format like JSON-LD (e.g. the dataset is an ontology), in which case it should still be listed under item, not as describedby.
Add a link relation for each of the metadata formats linked from the HTML, e.g.
<link
href="https://zenodo.org/records/7338056/export/json-ld"
rel="describedby" />Signposting recommends attributes for typed links, which are particularly important for metadata, which is available in many different formats.
The Bibliographic Metadata Formats listed for Signposting and the FAIR Signposting level 1 entry for
describedby lists common media types like application/vnd.datacite.datacite+xml
Augment the list of metadata resource to list the specific media types, e.g.:
<link
href="https://zenodo.org/records/7338056/export/bibtex"
rel="describedby"
type="application/x-bibtex"
/>Finally, some metadata use generic formats, like application/xml (XML) or application/ld+json (JSON-LD) -- the client will need to know particular namespaces or vocabularies to understand them. There may also be multiple metadata resources using the same format, but different models or variants. The concept of profile is intended for such disambiguating, and this is specified as an attribute of the link relation. The profile is identified by an URI.
To distinguish the two JSON-LD formats in this example, add their specific
profile:
<link
href="https://zenodo.org/records/7338056/export/json-ld"
rel="describedby"
type="application/ld+json"
profile="http://schema.org/"/>
<link
href="bioschemas.jsonld"
rel="describedby"
type="application/ld+json"
profile="https://bioschemas.org/profiles/Dataset/1.1-DRAFT"
/> For the Dublin Core export, add
profile="http://purl.org/dc/elements/1.1/" as suggested by the Bibliographic Metadata Formats table:
<link
href="https://zenodo.org/records/7338056/export/dublincore"
rel="describedby"
type="application/xml"
profile="http://purl.org/dc/elements/1.1/"/>While not required, it is good practice to link to the collection(s) the resource is from. In this case we don't have a persistent identifier for the collection.
Add the parent using
collection link relation:
<link
href="/signposting-tutorial/"
rel="collection">Finally, now let's consider the root index.html. The listing of datasets is not machine-readable.
A corresponding PID https://w3id.org/signposting-tutorial/USER should redirect to your dataset listing.
Modify
docs/index.html to add cite-as to reflect this persistent identifier:
<link
href="https://w3id.org/signposting-tutorial/stain"
rel="cite-as" />To indicate that this is a listing of datasets, use a type like https://schema.org/DataCatalog (for Dataset items), or https://schema.org/Collection (for any other types of items).
Modify
docs/index.html to add a type:
Note: We don't include AboutPage here, because the HTML listing is the collection.
Add the dataset using the link relation
item, but specify the type as text/html (items are landing pages):
<link href="7338056/"
rel="item"
type="text/html" /> For repositories there is likely to be very many entries. It is therefore NOT RECOMMENDED to list them in the HTTP headers, and they should not be listed in the HTML.
Remove the
item from before, and replace it with a linkset indirection:
<link href="linkset.json"
rel="linkset"
type="application/linkset+json" />A linkset is a mechanism to move links to a separate document . The document uses anchor to refer to which outgoing document the links are for, therefore a linkset can be common for multiple resources, which each link to it using linkset.
Inspect the docs/linkset.json for an example linkset in JSON.
Augment the
linkset.json file to include "type": "text/html" for the dataset 7338056. Remember to use correct , placement when editing the JSON.
{
"linkset": [
{
"anchor": "https://stain.github.io/signposting-tutorial/",
"cite-as": [
{
"href": "https://w3id.org/signposting-tutorial/stain"
}
],
"item": [
{
"href": "https://stain.github.io/signposting-tutorial/7338056/",
"type": "text/html"
}
]
}
]
} RFC9264 specifies the
application/linkset+json format, along with a text-based alternative application/linkset
In order to try out Signposting we will try two alternative Signposting clients.
Ensure you have committed and pushed your code to Github, allowing the page to rebuild. Visit the Actions tab in the GitHub repository to ensure the build succeeded as before.
After visiting your page, e.g. https://USER.github.io/signposting-tutorial/7338056/ , you may use Inspect Element in the Browser to check the <link> headers have been added correctly -- however your browser will by default not do any further validation.
If you are comfortable using the command line, and have Python 3.7 or later installed, then install the signposting Python package:
pip install signpostingVerify the tool is installed on the PATH:
(base) stain@xena:~/src/signposting-tutorial$ signposting --help
usage: signposting [-h] [--http] [--html] [--linkset] [-D] [-c] url [url ...]
positional arguments:
url URL(s) to discover signposting for
optional arguments:
-h, --help show this help message and exit
--http Find signposting in HTTP Link headers
--html Find signposting in <link> HTML elements if media-type matches
--linkset Find signposting in RFC9264 JSON or text linksets if media-type matches. When used with --recurse without specifying --http or --html, use those signposts to recurse, but
only report from linksets
-D, --distinct Report each signposting method (--http, --html and --linkset) separately
-c, --any-context Include signposts any contexts/anchors, not just resolved URINow try it on your GitHub deployment:
(base) stain@xena:~/src/signposting-tutorial$ signposting https://stain.github.io/signposting-tutorial/7338056/
Signposting for https://stain.github.io/signposting-tutorial/7338056/
CiteAs: <https://doi.org/10.5281/zenodo.7338056>
Type: <https://schema.org/AboutPage>
<https://schema.org/Dataset>
Collection: <https://stain.github.io/signposting-tutorial/>
License: <https://spdx.org/licenses/CC-BY-4.0>
Author: <https://orcid.org/0000-0003-2978-8922>
<https://orcid.org/0009-0004-1529-0095>
<https://orcid.org/0000-0003-3986-0510>
<https://orcid.org/0000-0002-1018-0370>
DescribedBy: <https://stain.github.io/signposting-tutorial/7338056/bioschemas.jsonld> application/ld+json
<https://zenodo.org/records/7338056/export/dublincore> application/xml
<https://zenodo.org/records/7338056/export/bibtex> application/x-bibtex
<https://zenodo.org/records/7338056/export/json-ld> application/ld+json
<https://zenodo.org/records/7338056/export/datacite-xml> application/vnd.datacite.datacite+xml
Item: <https://zenodo.org/records/7338056/files/Fleiss%20Kappa%20for%20document-to-document%20relevant%20assessment.csv?download=1> text/csvIf you have made a mistake, this library is likely to skip the particular signposting, or give a warning.
The Signposting Python library can also be used programmatically from other Python programs. See Signposting adopters for a complete list of software and repositories working with Signposting. alt text
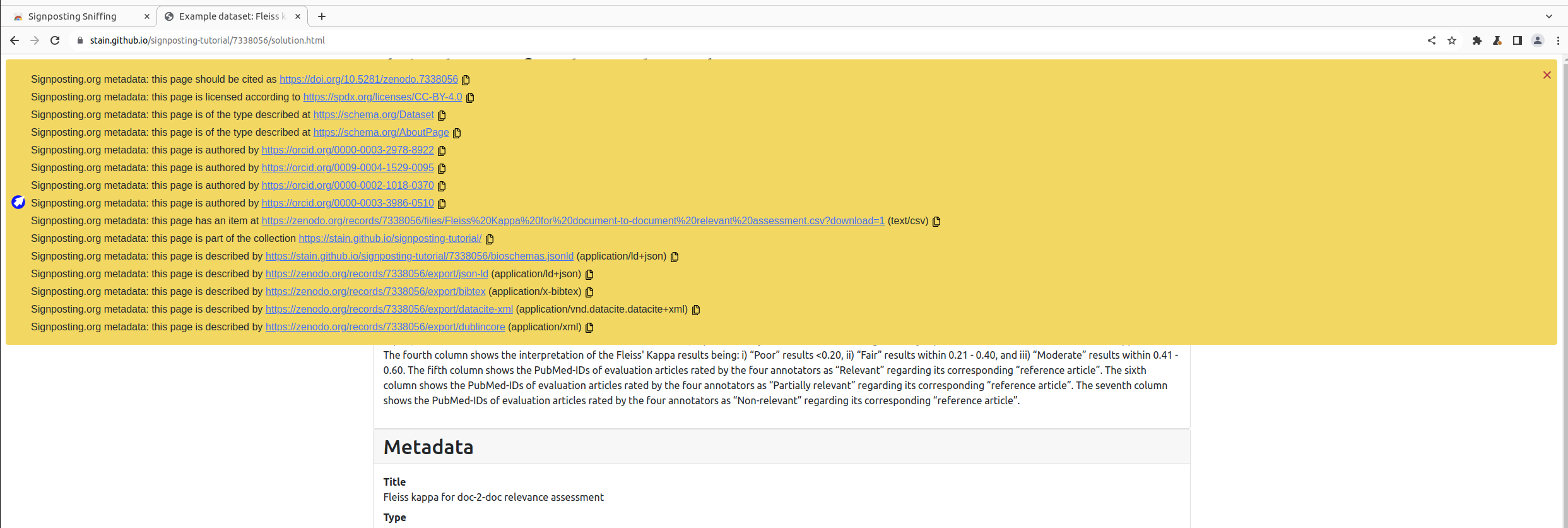
An experimental browser plugin for the Chrome browser (and its derivatives Chromium, Edge etc.) is available as Signposting Sniffing. Click Add to Chrome to enable this plugin. Note that although the plugin has access to inspect every web page, it should not be doing any external requests.
When Signposting is detected in a page, it will be presented as an overlay. After installing the plugin again, re-visit your dataset page in that browser.
This tutorial is based on bioschemas-ghpages-markup-tutorial, bioschemas-github-markup-example and Adding schema.org to a GitHub Pages site.
LJC has received fundings from the German Research Foundation (DFG) via the grant for NFDI4DataScience No. 460234259
We use free SVG icons from Font Awesone