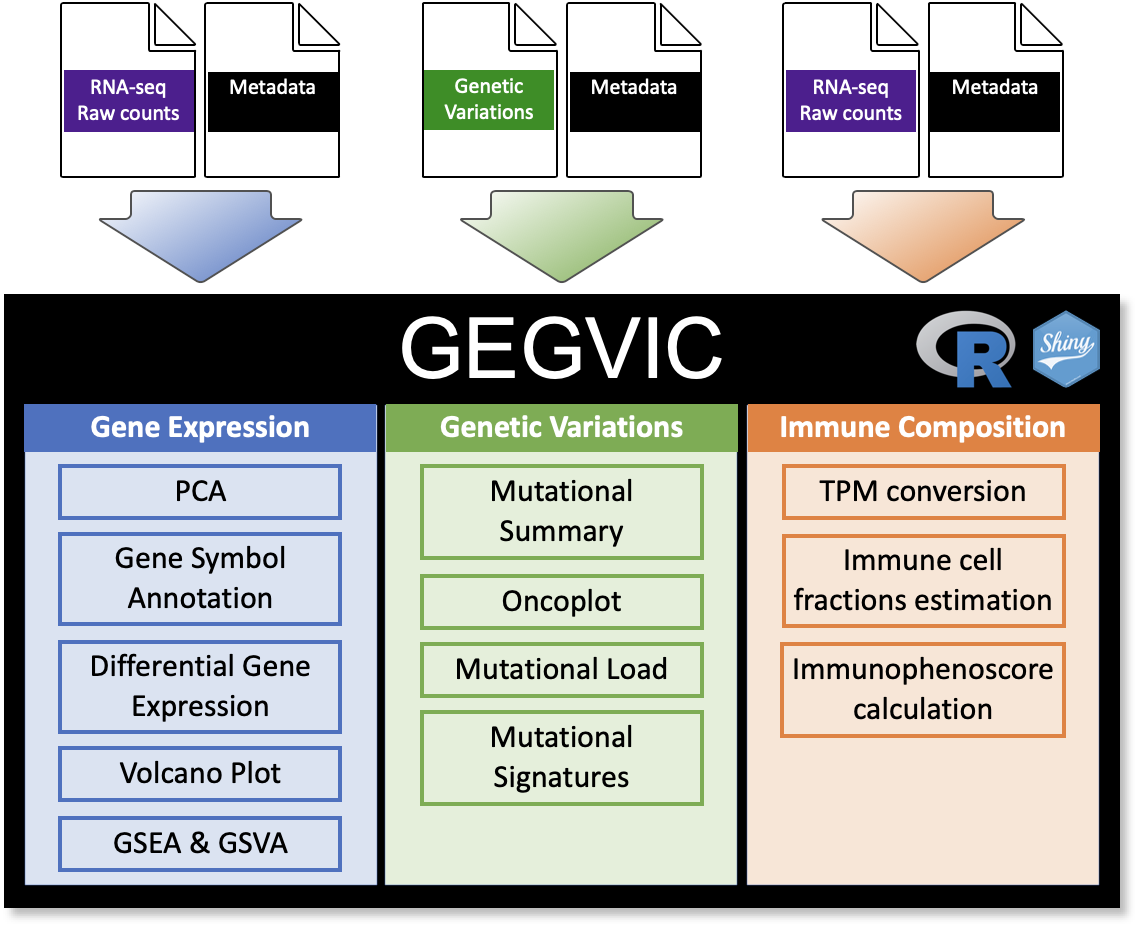
GEGVIC is a workflow to analyse Gene Expression, Genomic Variations and Immune cell Composition of tumour samples using Next Generation Sequencing data. This is a common need in the majority of the laboratories in the world, however, many times the high variety of tools available to perform each individual task can confuse and difficult the process.
Here we present an easy-to-use tool that requires few input files, provides a good flexibility and produces appealing outputs when comparing a group of samples for (i) differential gene expression, (ii) genomic variations and (iii) immune cell composition.
GEGVIC outlineYou can install the development version of GEGVIC from
GitHub with:
# install.packages("devtools")
devtools::install_github("oriolarques/GEGVIC")Since this package requires many dependencies, it is recommended to execute the following code before the first usage to prepare the environment correctly.
# CRAN packages
if(!require(shiny)) install.packages("shiny")
if(!require(dplyr)) install.packages("dplyr")
if(!require(tibble)) install.packages("tibble")
if(!require(tidyr)) install.packages("tidyr")
if(!require(ggplot2)) install.packages("ggplot2")
if(!require(ggrepel)) install.packages("ggrepel")
if(!require(rlang)) install.packages("rlang")
if(!require(ggplotify)) install.packages("ggplotify")
if(!require(ggpubr)) install.packages("ggpubr")
if(!require(patchwork)) install.packages("patchwork")
if(!require(gridExtra)) install.packages("gridExtra")
if(!require(pheatmap)) install.packages("pheatmap")
if(!require(devtools)) install.packages("devtools")
if(!require(remotes)) install.packages("remotes")
if(!require(DT)) install.packages("DT")
if(!require(shinyFiles)) install.packages("shinyFiles")
if(!require(shinythemes)) install.packages("shinythemes")
if(!require(tm)) install.packages("tm")
if(!require(rmarkdown)) install.packages("rmarkdown")
# Bioconductor packages
if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager")
if(!require(DESeq2)) BiocManager::install("DESeq2")
if(!require(apeglm)) BiocManager::install("apeglm")
if(!require(maftools)) BiocManager::install("maftools")
if(!require(clusterProfiler)) BiocManager::install("clusterProfiler")
if(!require(GSEAmining)) BiocManager::install("GSEAmining")
if(!require(GSEABase)) BiocManager::install("GSEABase")
if(!require(GSVA)) BiocManager::install("GSVA")
if(!require(SummarizedExperiment)) BiocManager::install("SummarizedExperiment")
if(!require(BSgenome)) BiocManager::install("BSgenome")
if(!require(BSgenome.Hsapiens.UCSC.hg19)) BiocManager::install("BSgenome.Hsapiens.UCSC.hg19")
if(!require(BSgenome.Hsapiens.UCSC.hg38)) BiocManager::install("BSgenome.Hsapiens.UCSC.hg38")
if(!require(BSgenome.Mmusculus.UCSC.mm10)) BiocManager::install("BSgenome.Mmusculus.UCSC.mm10")
if(!require(BSgenome.Mmusculus.UCSC.mm39)) BiocManager::install("BSgenome.Mmusculus.UCSC.mm39")
if(!require(DO.db)) BiocManager::install("DO.db")
if(!require(GO.db)) BiocManager::install("GO.db")
# Github packages
remotes::install_github("icbi-lab/immunedeconv")
devtools::install_github('raerose01/deconstructSigs')
devtools::install_github("oriolarques/GEGVIC")GEGVIC requires three main input data:
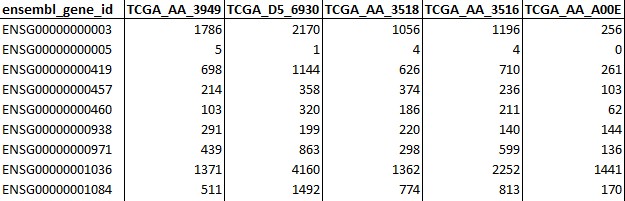
- RNA-sequencing raw counts (Counts): Table containing raw gene counts as rows and samples as columns. The first column must contain gene identifiers that can be either NCBI ID, ENSEMBL gene ID or HGNC ID and its column name MUST be adequately named as either:
-
entrezgene_id
-
ensembl_gene_id
-
hgnc_symbol
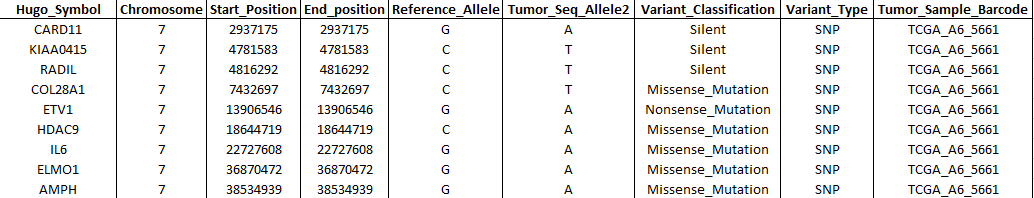
- Genomic variations data (Muts): Table containing short variant
calls. Necessary columns MUST have the following names
(following the MAF
format):
- Hugo_Symbol: Gene symbol from HGNC.
- Chromosome: Affected chromosome.
- Start_Position: Mutation start coordinate.
- End_Position: Mutation end coordinate.
- Reference_Allele: The plus strand reference allele at this position. Includes the deleted sequence for a deletion or “-” for an insertion.
- Tumor_Seq_Allele2: Tumor sequencing discovery allele.
- Variant_Classification: Translational effect of variant allele. Can be one of the following: Frame_Shift_Del, Frame_Shift_Ins, In_Frame_Del, In_Frame_Ins, Missense_Mutation, Nonsense_Mutation, Silent, Splice_Site, Translation_Start_Site, Nonstop_Mutation, RNA, Targeted_Region.
- Variant_Type: Type of mutation. Can be: ‘SNP’ (Single nucleotide polymorphism), ‘DNP’ (Double nucleotide polymorphism), ‘INS’ (Insertion), ‘DEL’ (Deletion).
- Tumor_Sample_Barcode: Sample name.
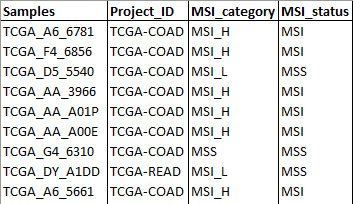
- Samples metadata: Table that contains additional information to the samples to create groups such as response to a therapy. The first column MUST be named Samples and contain the same nomenclature for each sample as in the RNA-sequencing raw counts and Genomic variations data tables.
Here, we will explore the workflow, how to use each function and the outputs that they generate. Users can use their own data (with the appropriate format as indicated before) by loading them in the R workspace, however, the package comes with pre-loaded input data from a subset of the TCGA-COADREAD cohort.
Notes:
- All the functions names have a prefix that indicate to which module they belong.
- For further information about specific function argument, install the GEGVIC package and use the help function or visit the description page for the corresponding function in the GitHub respository under the ‘R/’ section.
# load the package
library(GEGVIC)This module uses the functionalities provided by the DESeq2
package.
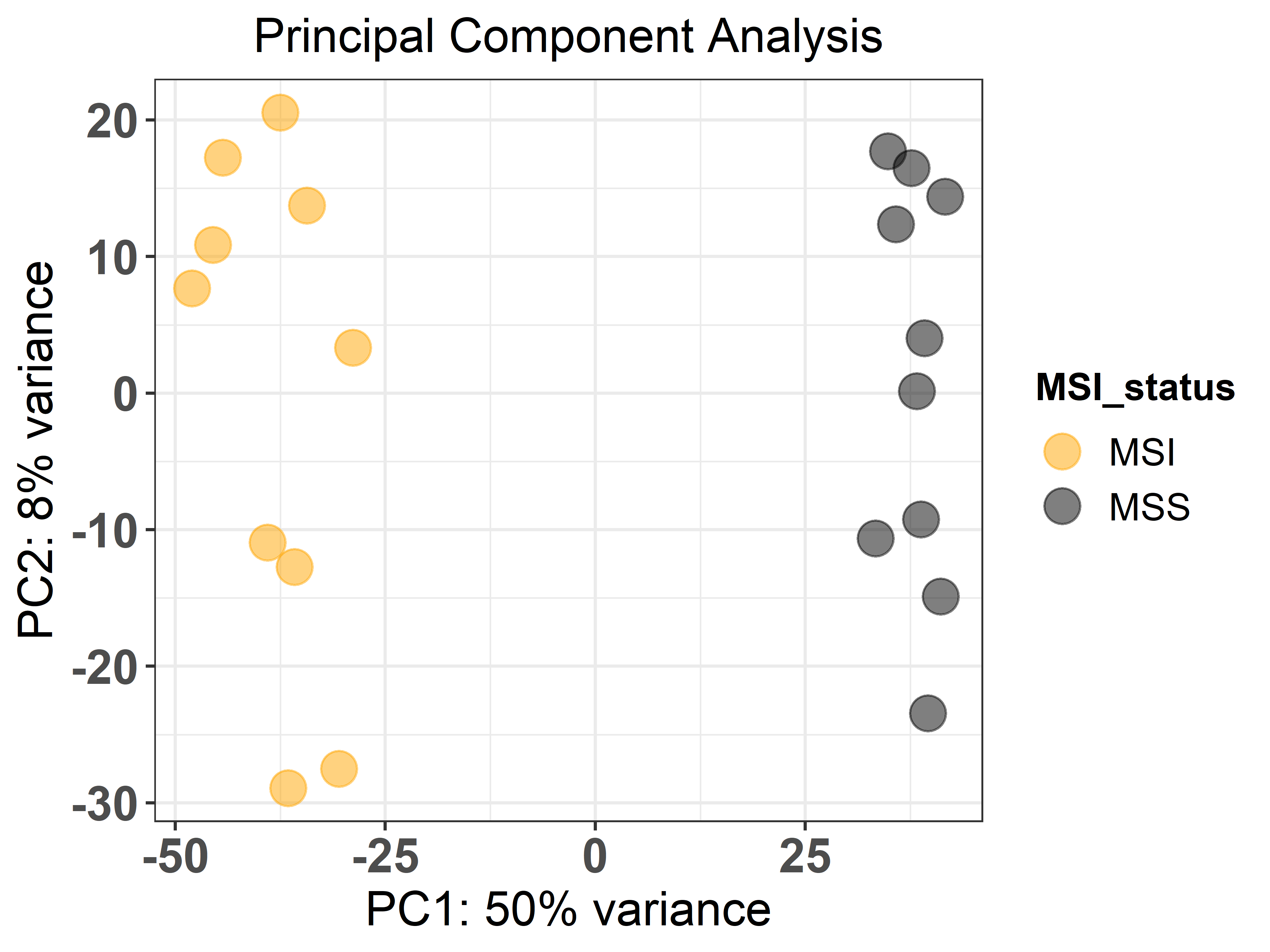
First, using the ge_pca() function we can perform a PCA to evaluate
how samples and groups relate to each other. For that, we indicate the
raw counts file (sample_counts), how the gene identifiers are encoded
(‘ensembl_gene_id’), the metadata file (sample_metadata) and the
unquoted name of the column that contains the groups of interest as
the response argument. Then, the design should be a formula that
expresses how the counts for each gene depend on the variables in the
metadata, and finally the colours to represent each sample group. The
function outputs a plot.
ge_pca(counts = sample_counts,
genes_id = 'ensembl_gene_id',
metadata = sample_metadata,
response = MSI_status,
design = 'MSI_status',
colors = c('orange', 'black'))Then, we can compute differential gene expression between groups of
interest using the ge_diff_exp() function and store the results in an
object (results.dds).
We need to define new parameters such as the samples group that will be used as the level of reference (the group to which the others will be compared against in a form of a vector) and the shrinkage method of the log2 fold changes to be applied (or not).
In the case there are multiple levels of comparison the object will be in a form of a list.
results.dds <- ge_diff_exp(counts = sample_counts,
genes_id = 'ensembl_gene_id',
metadata = sample_metadata,
design = 'MSI_status',
ref_level = c('MSI_status', 'MSS'),
shrink = 'apeglm')In the case that the gene identifiers provided are not in form of HGNC
symbols but are NCBI or ENSEMBL ID, we have to use the ge_annot()
function to perform the appropriate conversion and store the results in
a new object (annot.res). For that we will have to indicate a query
from the biomart
package
with the following attributes: ensembl_gene_id, hgnc_symbol,
entrezgene_id, transcript_length, refseq_mrna. GEGVIC has already
available the following databases:
-
Genome Reference Consortium Human Build 37: ensembl_biomart_GRCh37.
-
Genome Reference Consortium Human Build 38: ensembl_biomart_GRCh38_p13.
-
Genome Reference Consortium Mouse Build 38 (mm10): ensembl_biomart_GRCm38_p6.
-
Genome Reference Consortium Mouse Build 39 (mm39): ensembl_biomart_GRCm39.
annot.res <- ge_annot(results_dds = results.dds,
genes_id = 'ensembl_gene_id',
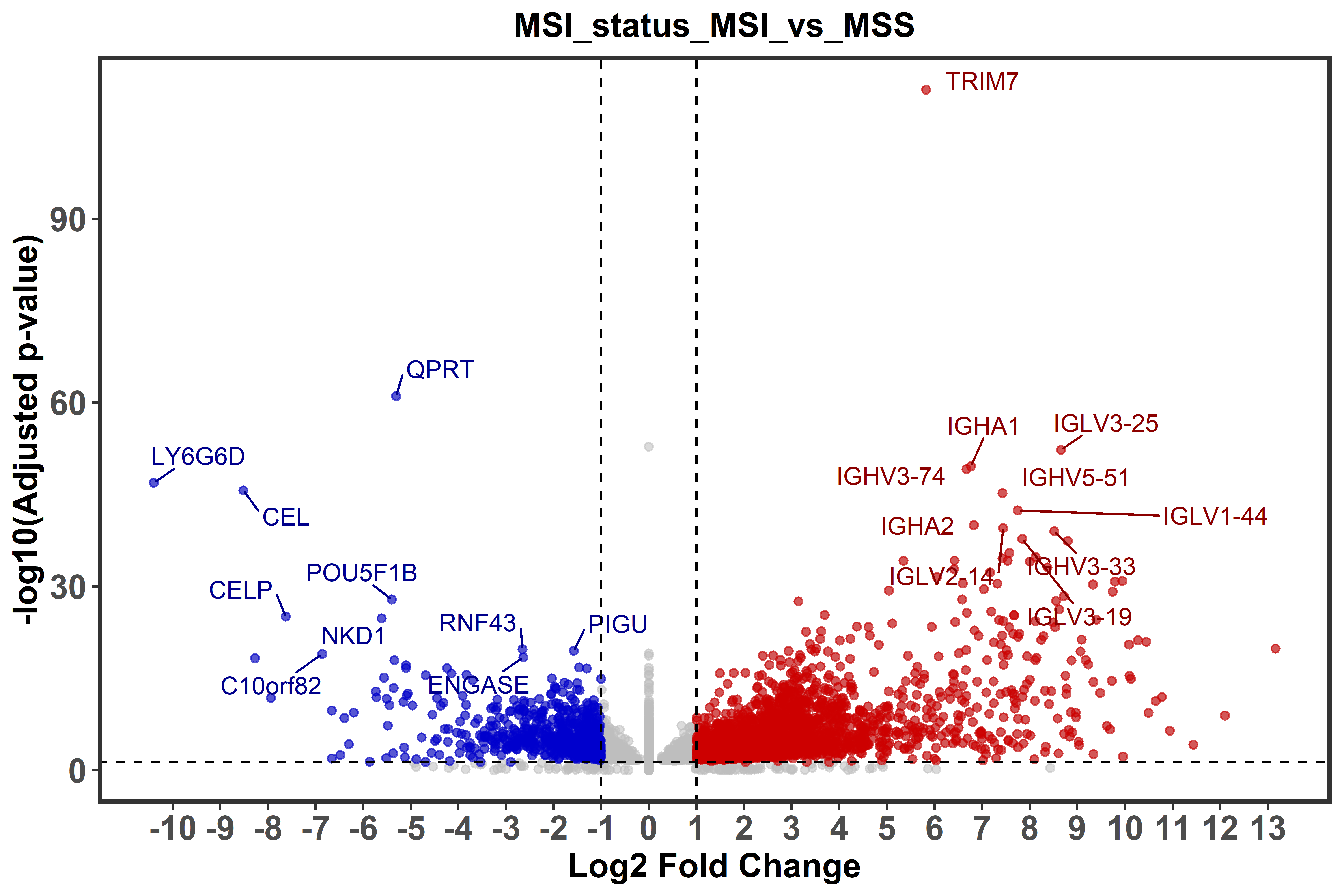
biomart = ensembl_biomart_GRCh38_p13)To represent differential gene expression in form of Volcano plots the
function ge_volcano() is used to generate a plot for each comparison
groups. In the plot, the top ten most significantly up- and dw-regulated
genes will be highlighted. Furthermore, the function allow users to
define the fold change and adjusted p-value to further customize the
plot.
ge_volcano(annot_res = annot.res,
fold_change = 2,
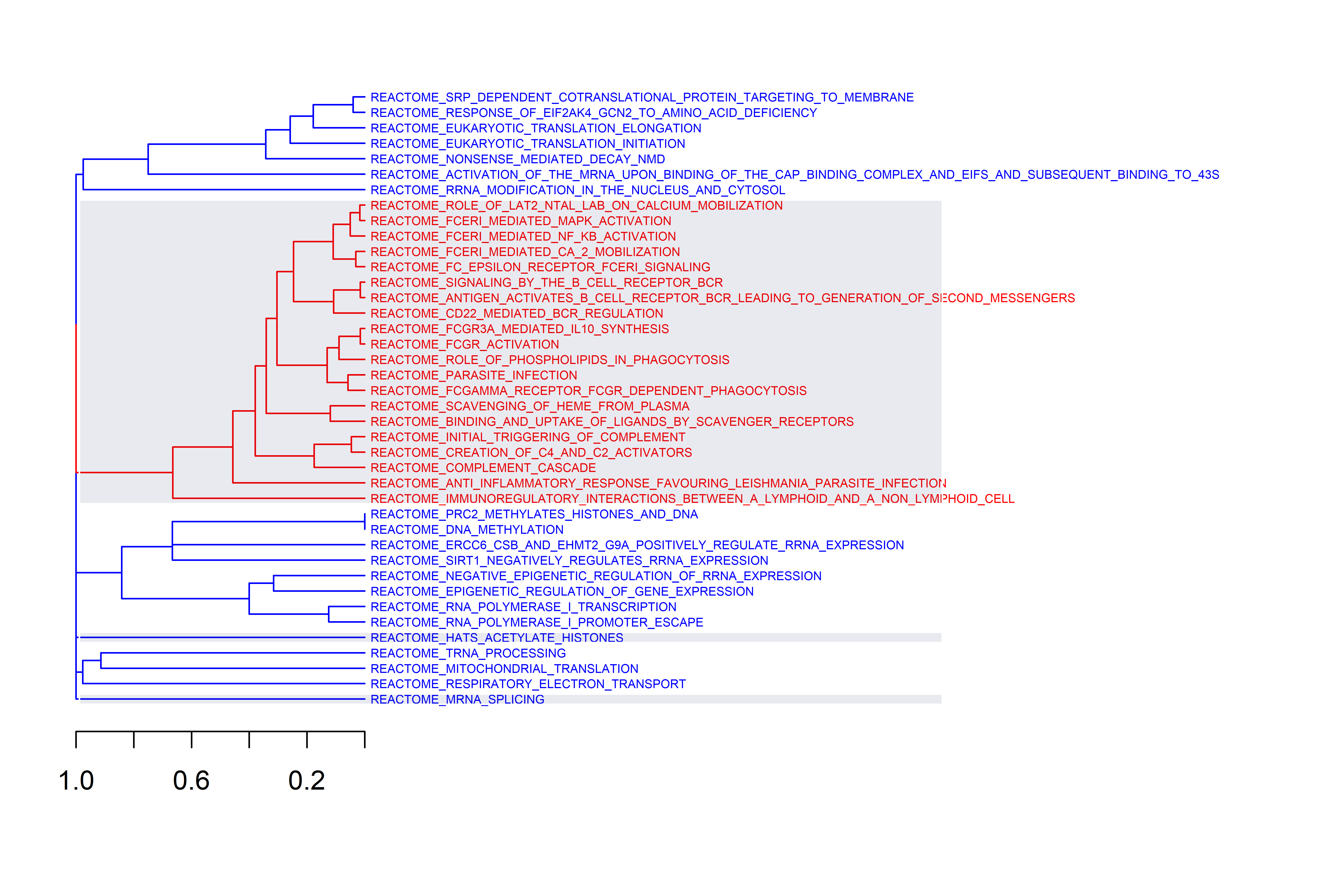
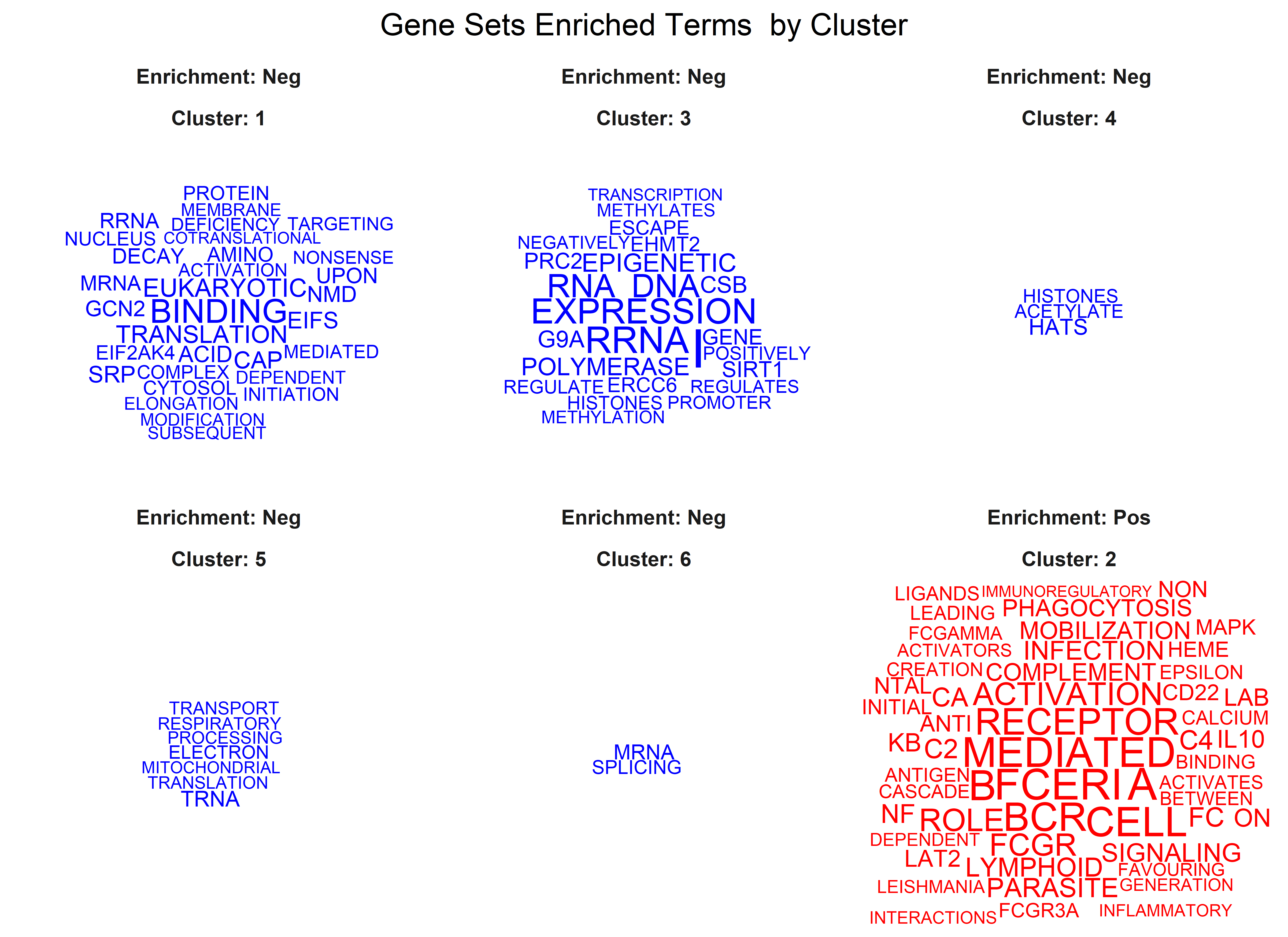
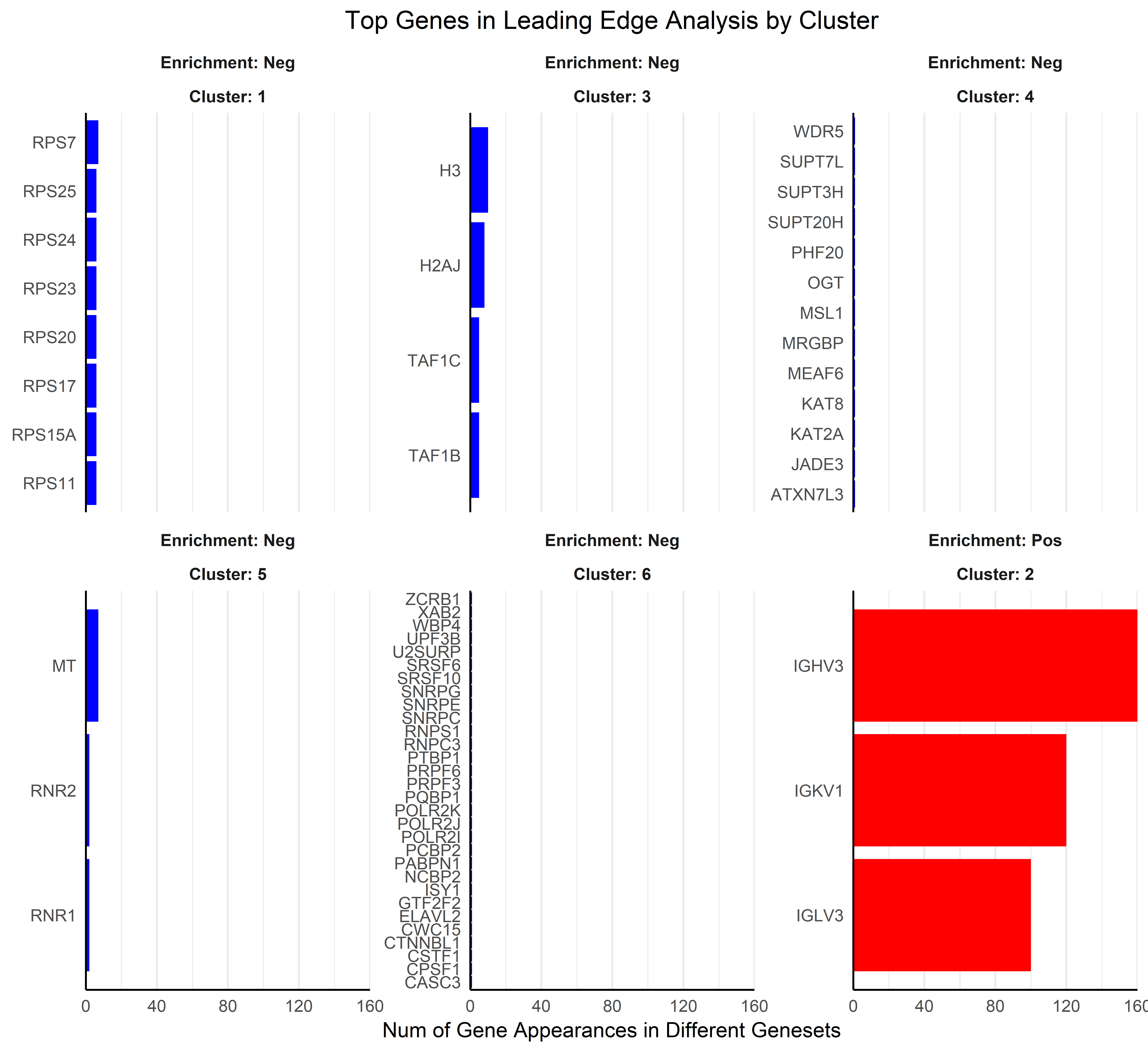
p.adj = 0.05)One of the last functions of the module, ge_gsea(), permits to perform
Gene Set Enrichment Analysis (GSEA) using the clusterProfiler
package
functionalities. The resulting top 20 regulated gene sets are shown in a
bubble plot where Normalized Enrichment Score (NES) is shown. The size
of the bubbles are determined by the percentage of genes in the gene set
that belong to the leading edge (core). Then, the same gene sets are
grouped by similarity and plotted using the GSEAmining
package.
Three plots are generated, first a cluster of gene sets and, per each
cluster, a wordcloud of biological terms enriched in each case and the
top 3 genes in the leading edge of the different gene sets present in
that cluster.
To use this function the user has to provide a collection of gene sets to evaluate in a form of a gmt file. This can be downloaded from the Molecular Signatures Database, MSigDB or be customly created following the corresponding [guidelines] (https://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/Data_formats). In the case of working with mouse data, gene symbols are automatically transformed to human orthologs, so the same gene sets from MSigDB can be used. The Reactome gene sets were downloaded (c2.cp.reactome.v7.5.1.symbols.gmt file) and used to create this example.
Additionally, users can define the adjusted p-value cut-off to be more or less restrictive when performing GSEA. The function generates two plots, one dendrogram and one wordcloud with the most enriched name terms in each cluster in the dendrogram.
Note: There are two ways to access to the results table. (1) Call the
object results object as gsea.res$table_name@result or (2)
as.data.frame(gsea.res).
gsea.res <- ge_gsea(annot_res = annot.res,
gmt = 'inst/extdata/c2.cp.reactome.v7.5.1.symbols.gmt',
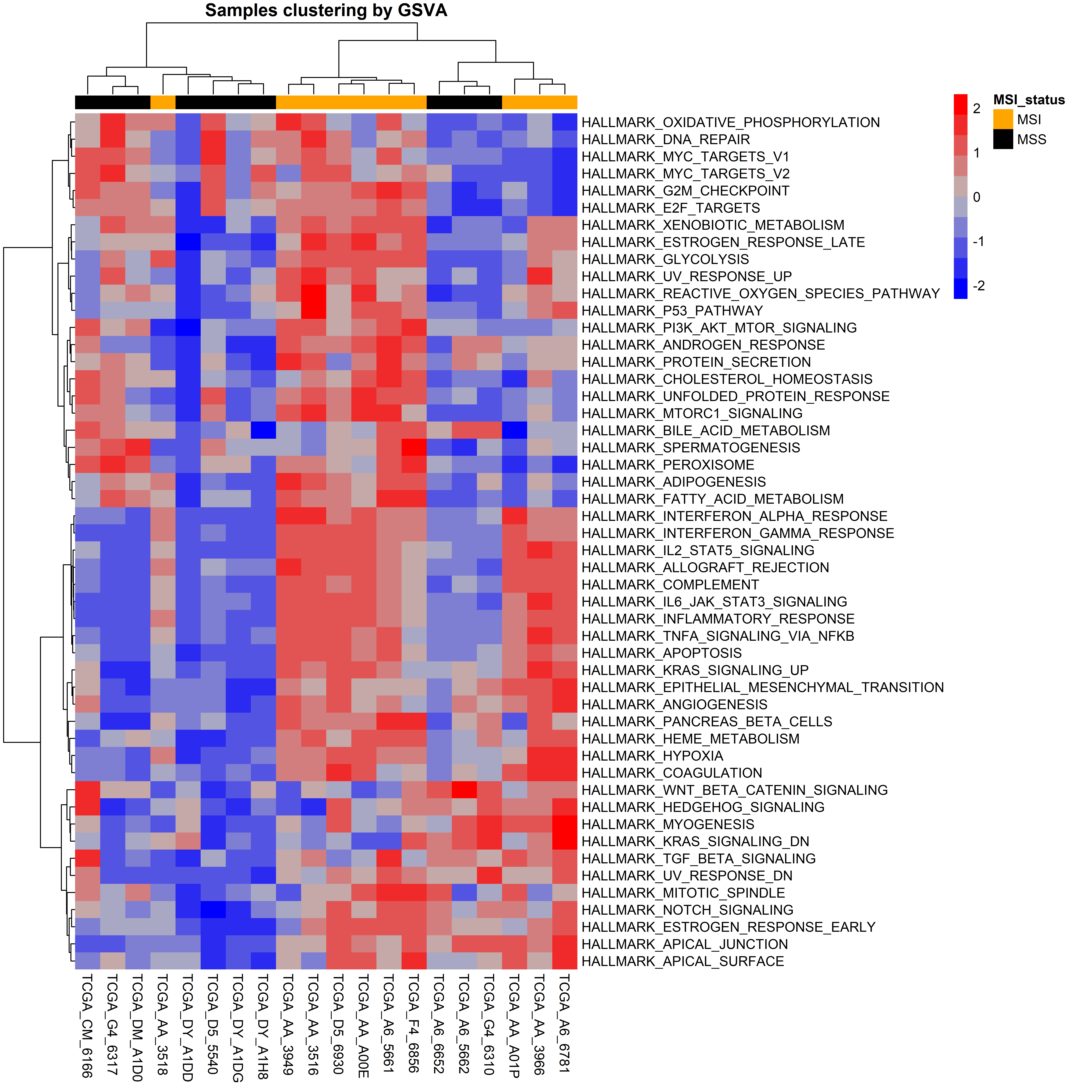
gsea_pvalue = 0.2)Finally, the ge_single() allows to perform Gene Set Variation Analysis
(GSVA) or single sample GSEA (ssGSEA) to cluster samples using the
GSVA
package.
In order to do that, the user has to define the method and also indicate
the gene set collection of interest. By default, the HALLMARK
collection from the MSigDB will be used if the location of a different
.gmt file is not provided.
Results are shown as a heatmap. Users can define the color of the sample groups and also if gene set names and/or sample names should be plotted or not.
gsva.res <- ge_single(counts = sample_counts,
metadata = sample_metadata,
genes_id = 'ensembl_gene_id',
response = MSI_status,
design = 'MSI_status',
biomart = ensembl_biomart_GRCh38_p13,
gsva_gmt = 'hallmark',
method = 'gsva',
kcdf = 'Gaussian',
colors = c('orange', 'black'),
row.names = TRUE,
col.names = TRUE)This module uses functionalities from the immunedeconv
package.
To predict immune composition of tumour microenvironment from
RNA-sequencing data, we need first to transform raw counts to TPM
(Transcript Per kilobase Million), as it is required by all the methods
in the immunedeconv package. The function ic_raw_to_tpm() takes the
same input as for the GE_module (RNA-seq raw counts). It also needs the
gene identifiers encoding and the biomaRt database. The results should
be stored as a new object (i.e.: tmp).
tpm <- ic_raw_to_tpm(counts = sample_counts,
genes_id = 'ensembl_gene_id',
biomart = ensembl_biomart_GRCh38_p13)The object containing TPM reads will be used as the input for the
ic_deconv() function, which estimates the immune cell composition of
the samples. All the following methods (included in the immunedeconv
package) QUANTISEQ, TIMER, MCP_COUNTER, XCELL, EPIC and CIBERSORT will
be used.
Note: To use the CIBERSORT, the user need to register on the CIBERSORT web page (https://cibersort.stanford.edu), obtain a license and download the source code in form of two files CIBERSORT.R and LM22.txt. Then, the user need to specify the path to the storage location of such files in the cibersort argument.
The indications argument must be a character vector of cancer type codes for each sample in the tpm matrix. Indications supported can be checked using immunedeconv::timer_available_cancers. Results should be saved in a new object (i.e.: ic.pred).
ic.pred <- ic_deconv(gene_expression = tpm,
indications = rep('coad', ncol(tpm)),
cibersort = 'cibersort/', # Set to NULL to not use this option
tumor = TRUE,
rmgenes = NULL,
scale_mrna = TRUE,
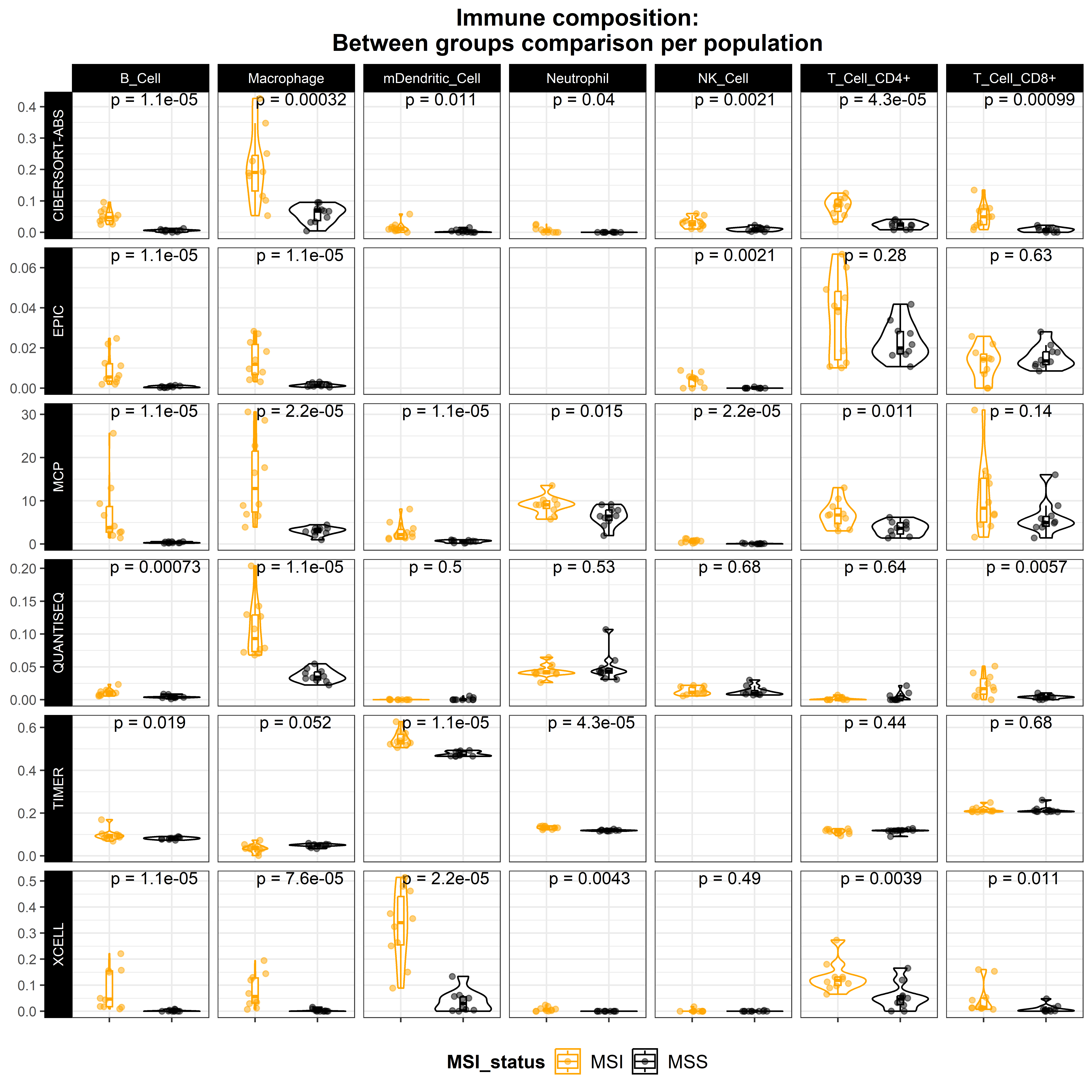
expected_cell_types = NULL)With the ic_plot_comp_samples() function we can plot a graph comparing
each immune cell populations between sample groups per method. For that,
the name of column where lies the grouping variable must be written
WITHOUT quotes in the response argument. The compare argument allow
users to decide which method should be used for comparing means. Options
are ‘t.test’ and ‘wilcox.test’ for two groups or ‘anova’ and
‘kruskal.test’ for more groups. Also, the p_label argument permits to
choose the way the significance is represented, being either ‘p.signif’
(shows the significance levels) or ‘p.format’ (shows the formatted
p-value). The function allows to change also the colours of the groups
and decide if points are added to the plot.
ic_plot_comp_samples(df = ic.pred,
metadata = sample_metadata,
response = MSI_status,
compare = 'wilcox.test',
p_label = 'p.format',
colors = c('orange', 'black'),
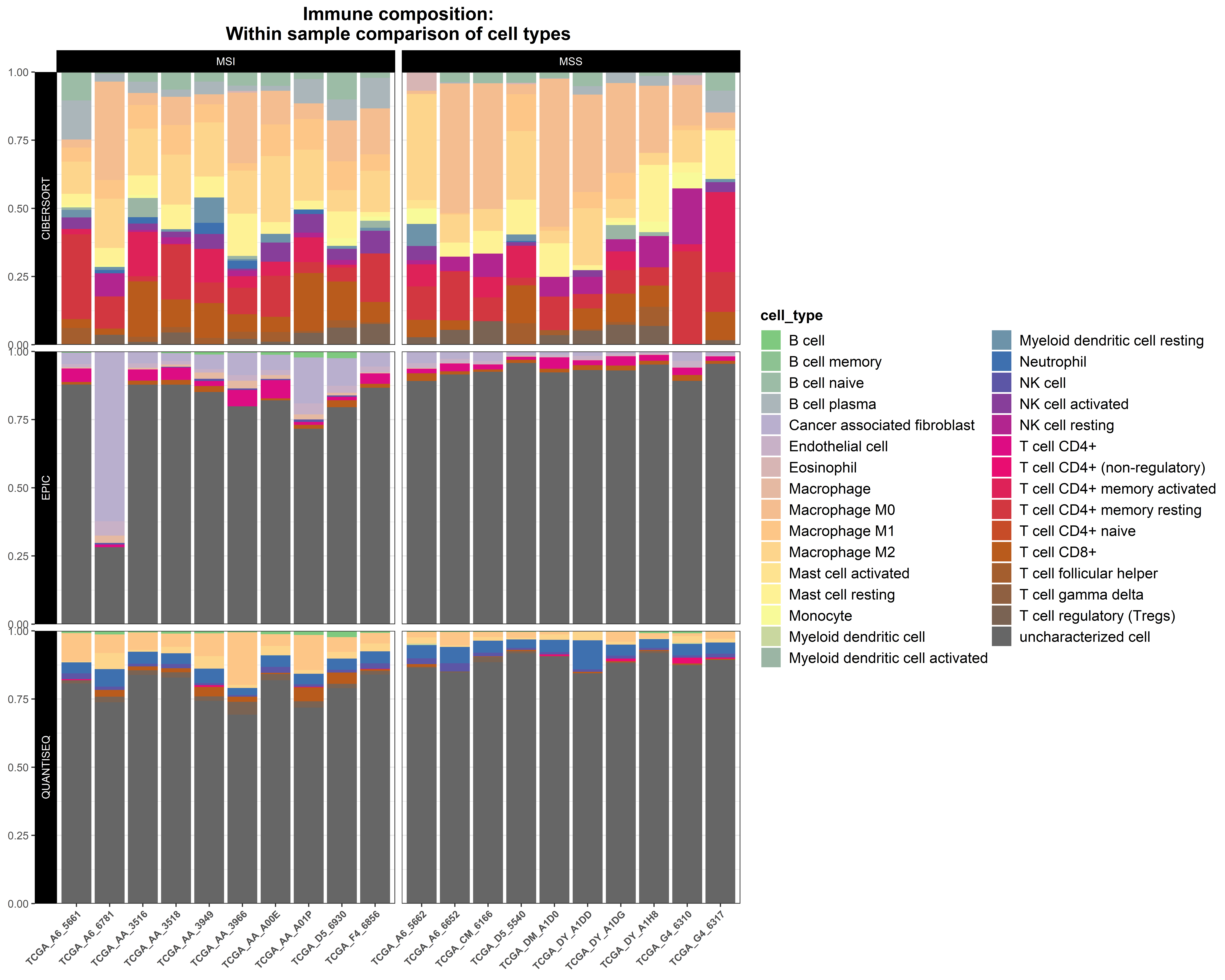
points = FALSE)Similarly, the ic_plot_comp_celltypes() function is able to plot the
comparison of each immune cell fraction within each sample from the
predictions made by CIBERSORT, EPIC and QUANTISEQ.
ic_plot_comp_celltypes(df = ic.pred,
metadata = sample_metadata,
response = MSI_status,
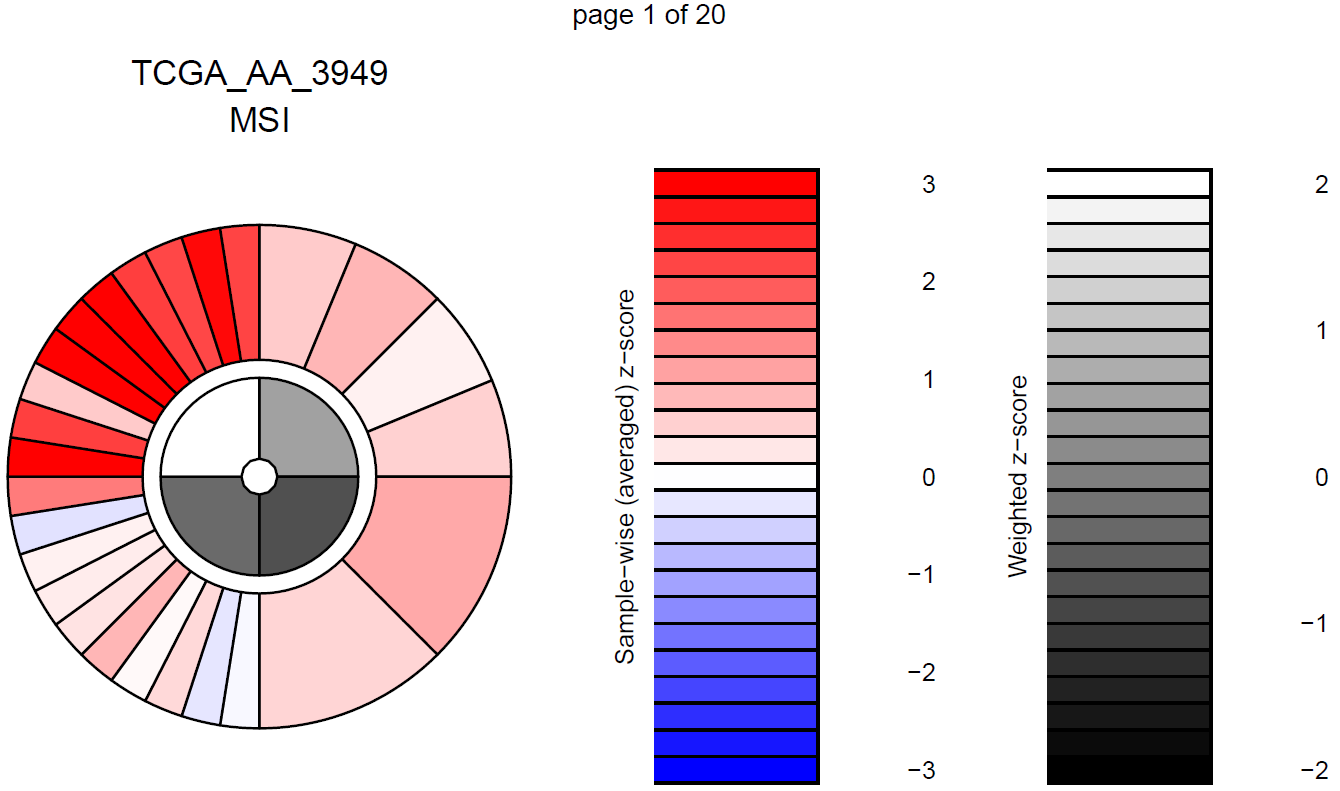
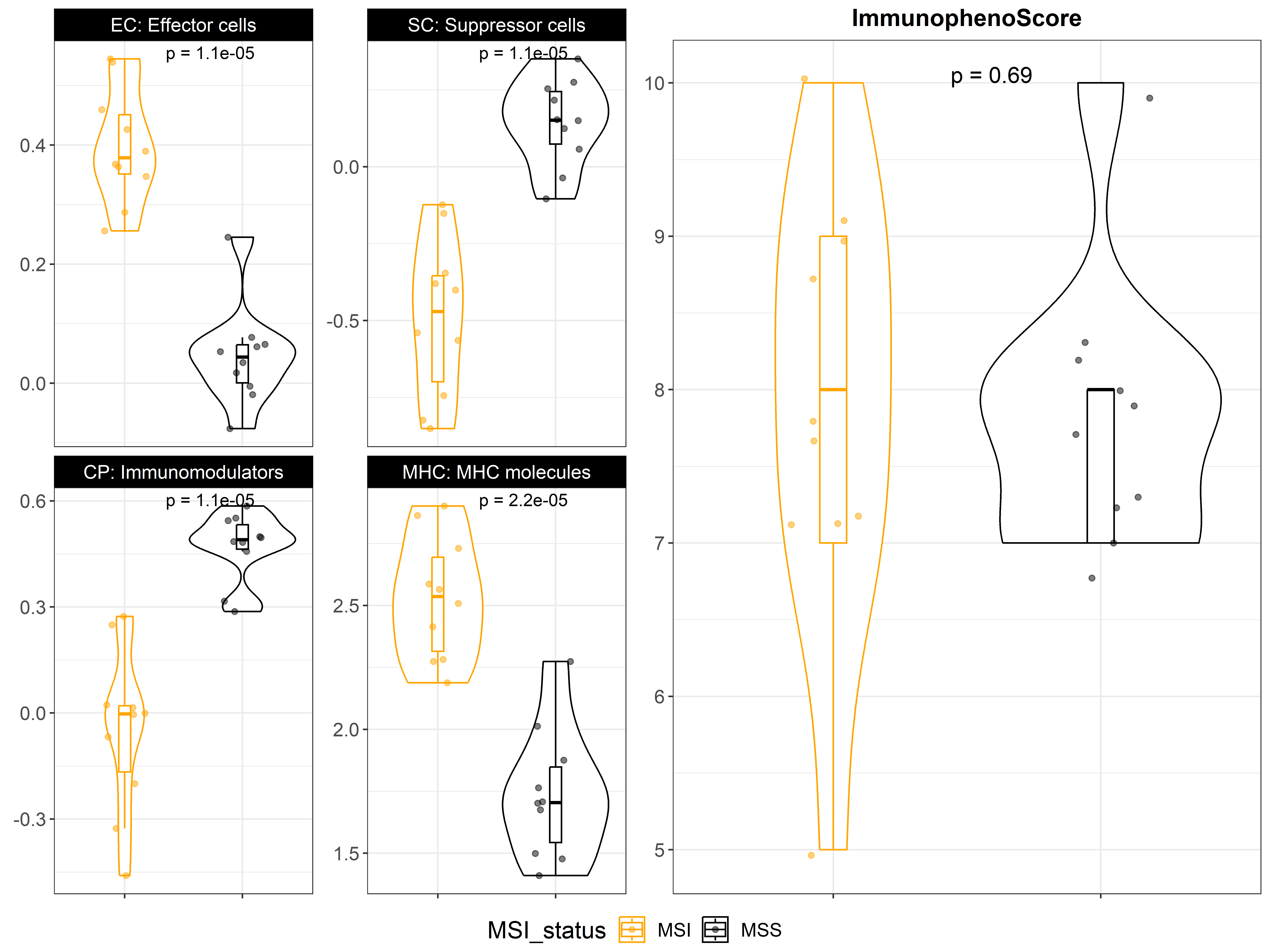
col.names = TRUE)The last function in this module, ic_score() uses TPM expression
values to calculate and plot immunophenogram (IPG) and immunophenoscores
(IPS) for each sample and each group of study. They give an overall
picture of the state of MHC molecules (MHC), Immunomodulators (CP),
Effector cells (EC) and Suppressor cells (SC) in each sample, making
possible the comparison between samples. For further interpretation
please visit https://tcia.at/tools/toolsMain.
Note: Immunophenograms are generated in a pdf file containing the
results for each sample in a single page. The output file, named
immunophenogram_report.pdf, will be stored in the working
directory.
ips <- ic_score(tpm = tpm,
metadata = sample_metadata,
response = MSI_status,
compare = 'wilcox.test',
p_label = 'p.format',
colors = c('orange', 'black'))This module uses functionalities from the maftools
package
and the deconstructSigs
package.
The genomic variations input (sample_mutations) together with samples
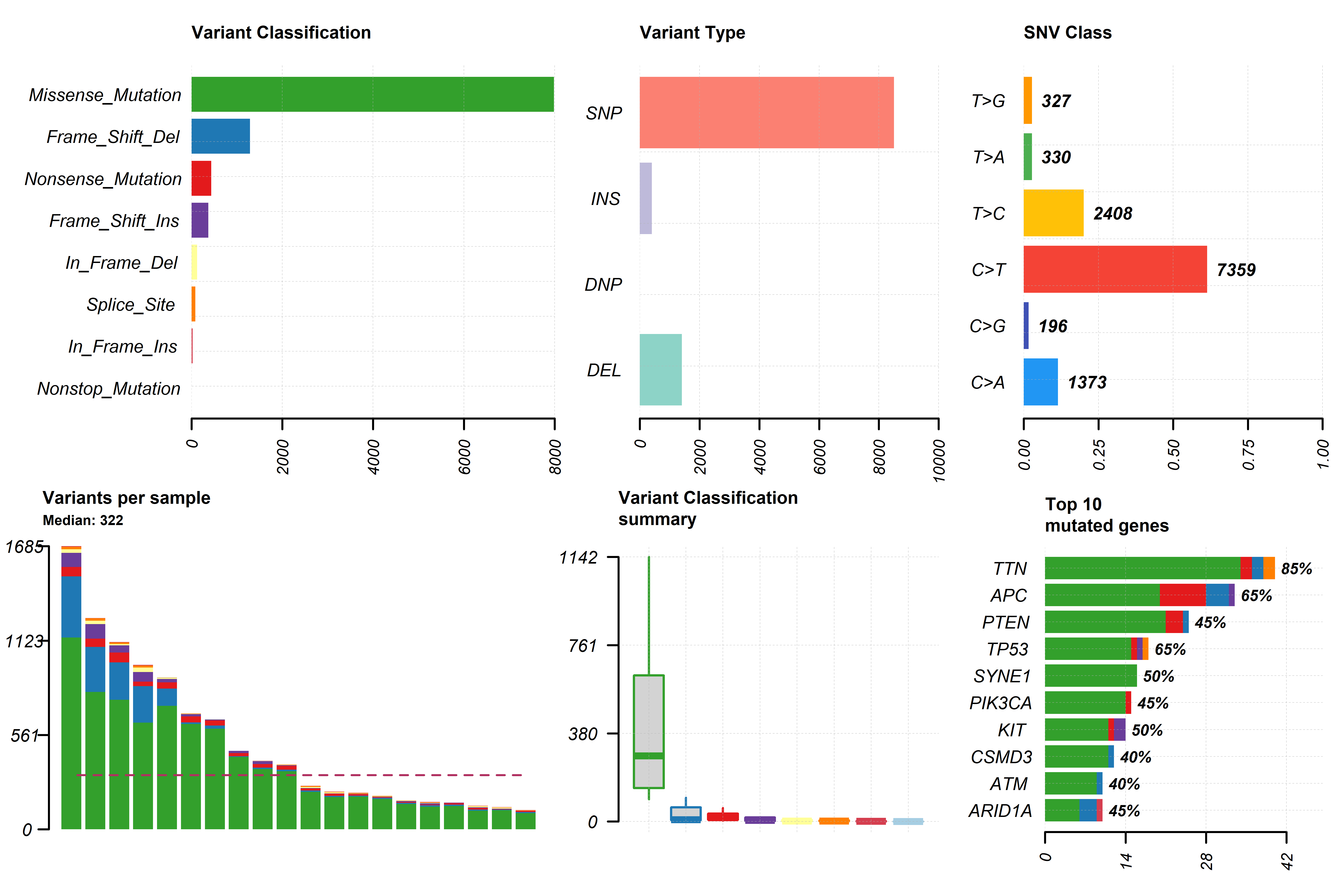
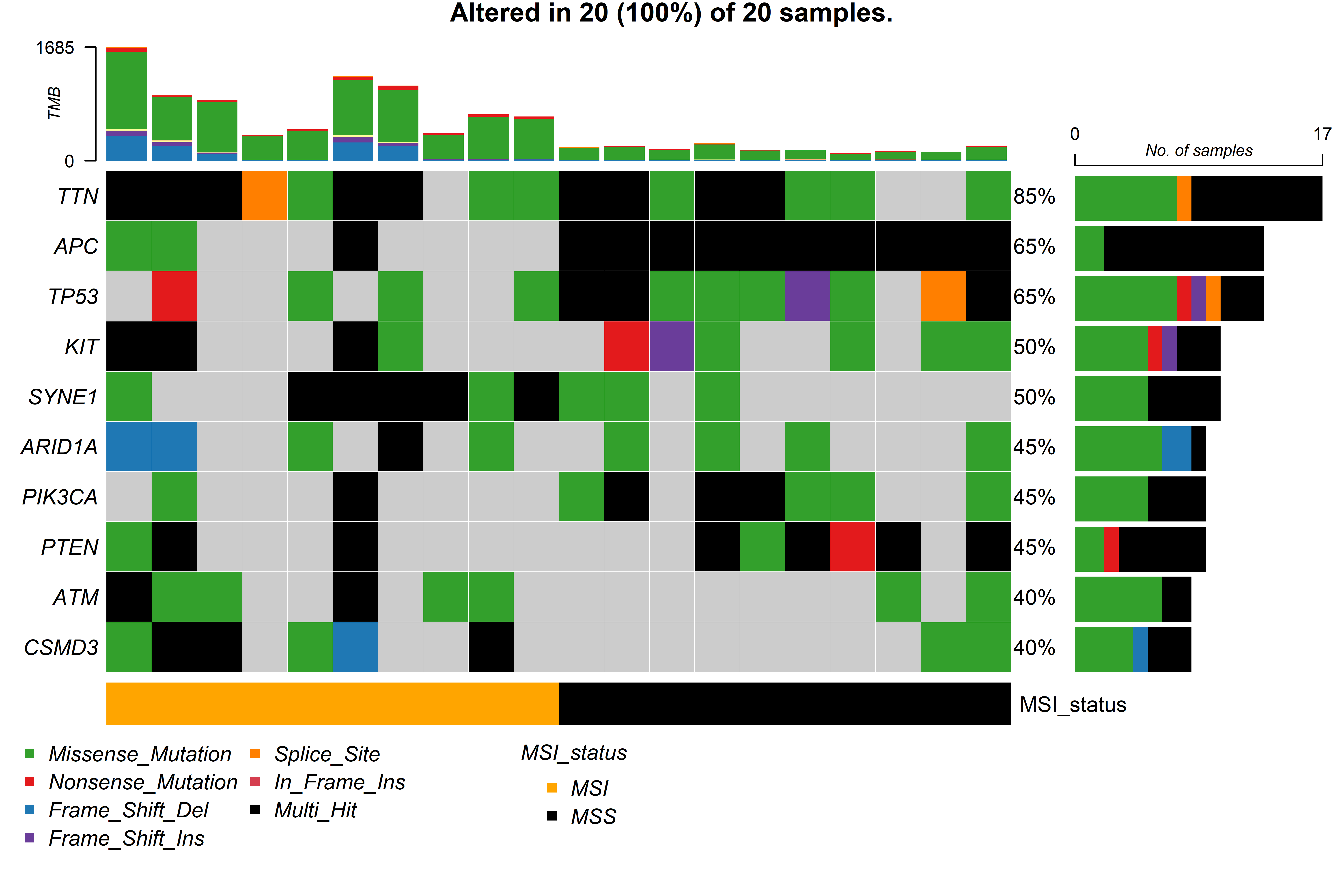
metadata can be used in the function gv_mut_summary() to generate two
plots that will first summarise the mutation types present in the
samples and second highlight the most common mutations by groups in a
form of an oncoplot.
Users MUST indicate the unquoted name of the column that contains the groups of interest in the response argument. Additionally, parameters that define the number and which genes will appear in the oncoplot, the colours of sample groups and whether the names of the samples will appear in the plot can be modified.
gv_mut_summary(muts = sample_mutations,
metadata = sample_metadata,
response = MSI_status,
top_genes = 10,
specific_genes = NULL,
col.names = FALSE,
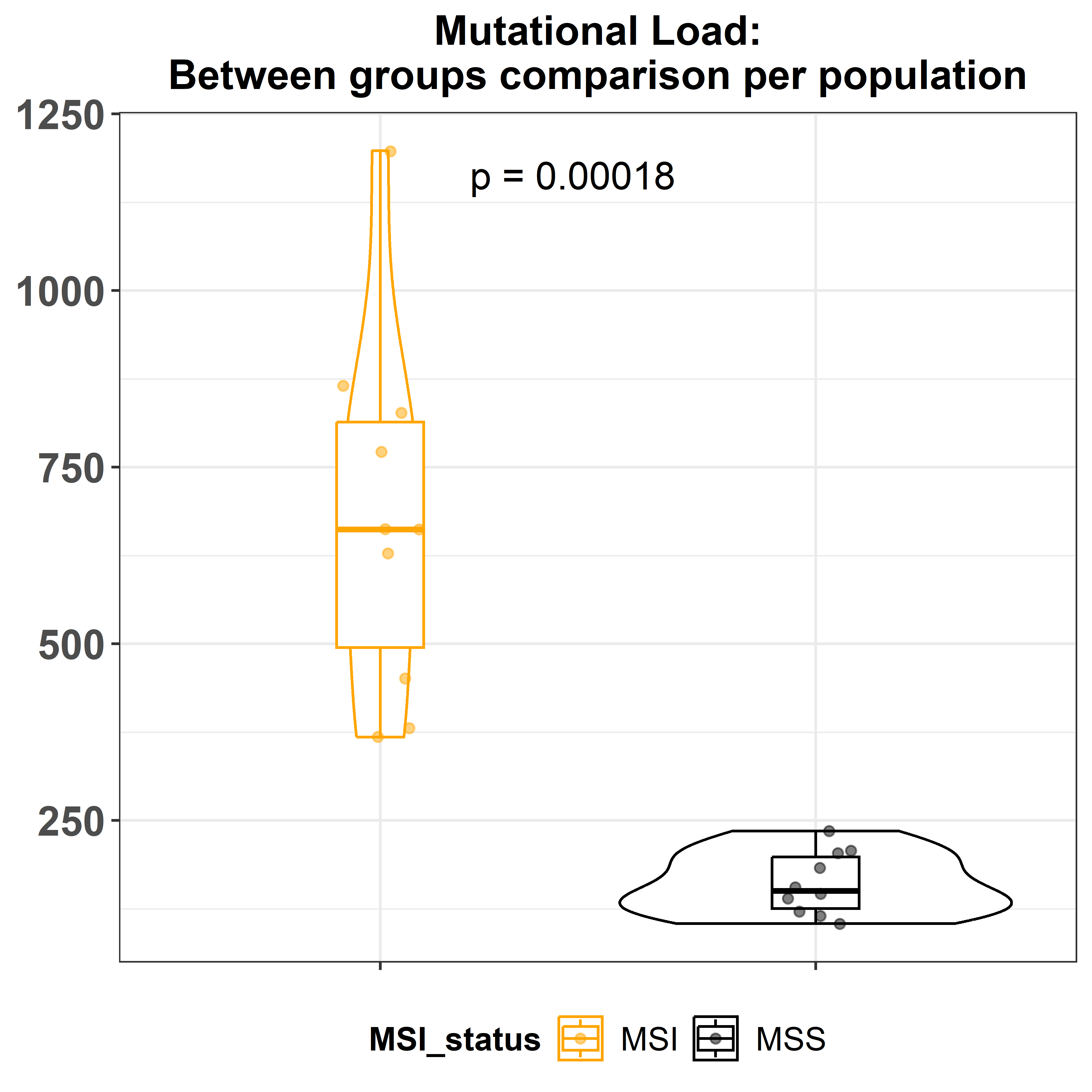
colors = c('orange', 'black'))The function gv_mut_load() will calculate the total number of
mutations per sample. The same inputs as the previous function are
required. Also the compare and p_label arguments allow users to decide
which method should be used for comparing means and the way the
significance is represented. As usual, the function allows to change
also the colours of the groups.
mut.load <- gv_mut_load(muts = sample_mutations,
metadata = sample_metadata,
response = MSI_status,
compare = 'wilcox.test',
p_label = 'p.format',
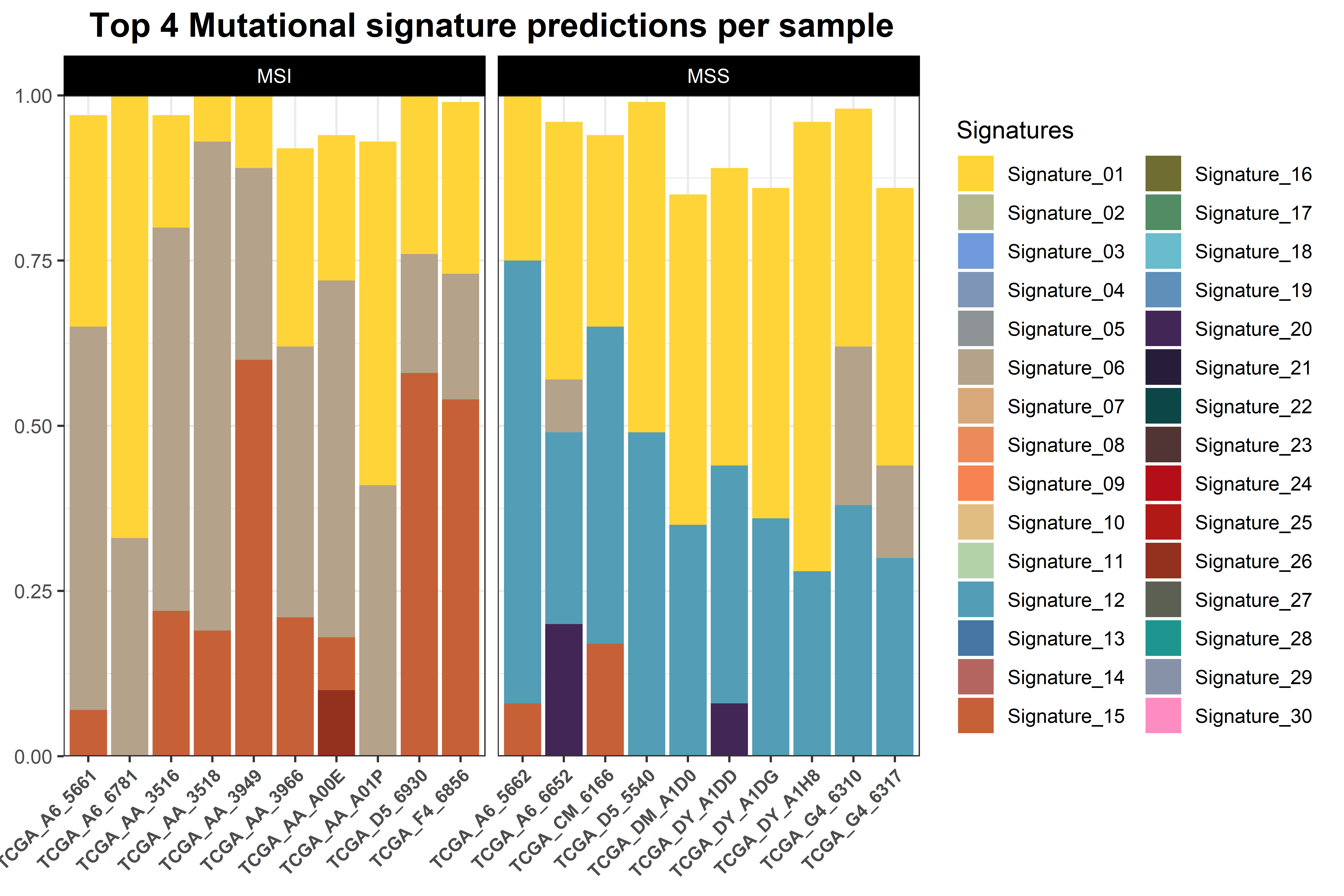
colors = c('orange', 'black'))The last function of the module,gv_mut_signatures() is used to predict
the weight of mutational signatures contributing to an individual tumour
sample. As well as the inputs described before, here the user have to
choose the version of the genome to work with. To do so, the gbuild
argument should be one of the following:
-
‘BSgenome.Hsapiens.UCSC.hg19’
-
‘BSgenome.Hsapiens.UCSC.hg38’
-
‘BSgenome.Mmusculus.UCSC.mm10’
-
‘BSgenome.Mmusculus.UCSC.mm39’
Also, the mutational signature matrices containing the frequencies of all nucleotide changes per signature need to be indicated. GEGVIC contains the matrices from COSMIC for single and double base substitutions.
To choose one, the user has to indicate ’COSMIC_v{XX}_{YY}BS_GRCh{ZZ}’ (i.e. ‘COSMIC_v2_SBS_GRCh37’) in the mut_sigs argument:
-
XX is the version, that can be v2 or v3.2.
-
YY indicates if mutations are single (S) or double (D) base substitutions.
-
ZZ is for the genome assembly, either GRCh37 or GRCh38 for human data and mm9 or mm10 for mouse data.
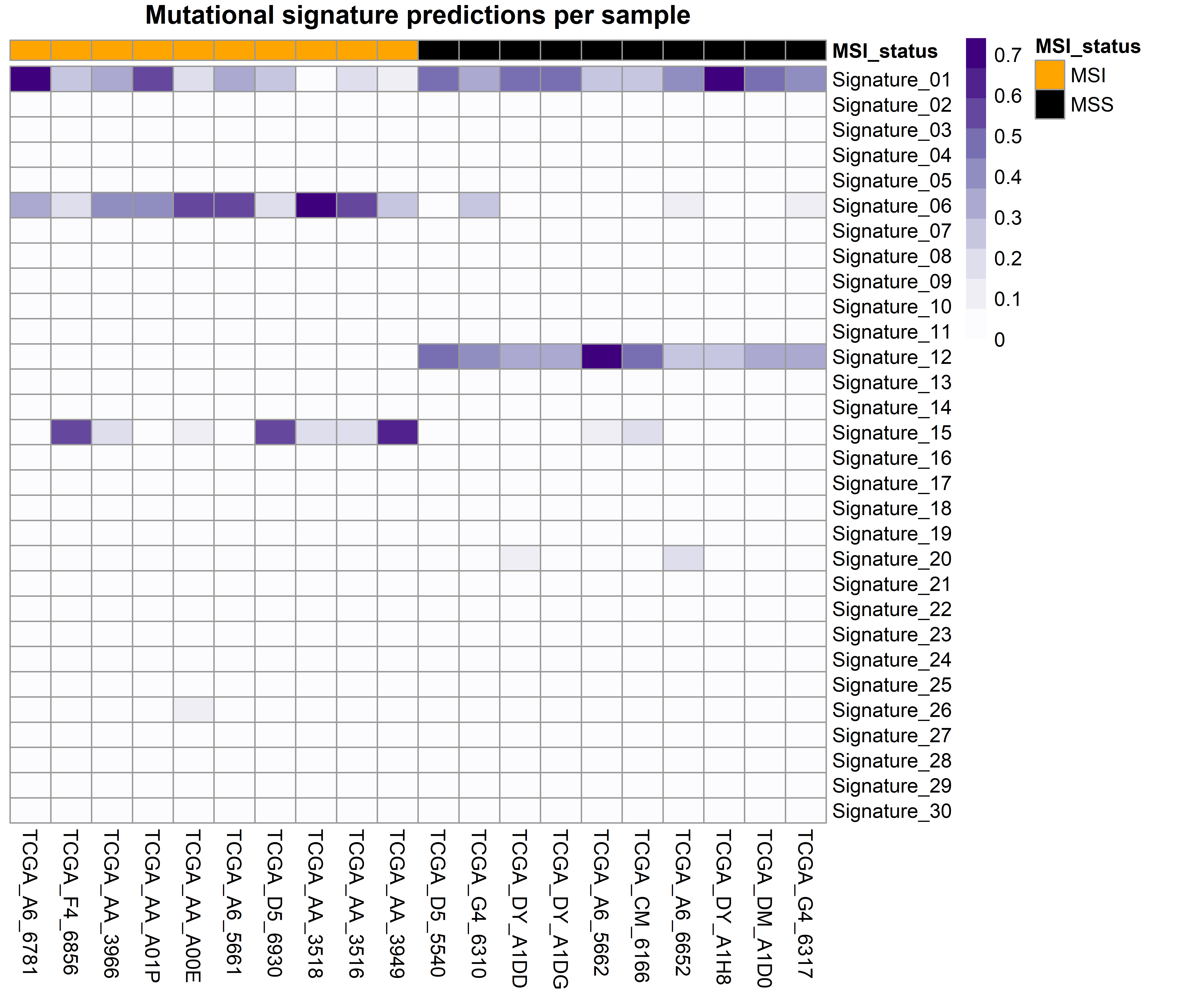
The function generates two plots. The first is a barplot that shows the weight of the top four mutational signatures per sample and group. Since depending on how many samples or signatures are present in the analysis the results may be difficult to interpret, a second plot is generated. This is a heatmap that shows all samples as columns and signatures as rows and the weight of each signature determines the intensity of the colour. We believe that, although the first plot is more common in the literature, the second plot can be helpful, especially when many mutational signatures are present in many samples.
mut.sigs <- gv_mut_signatures(muts = sample_mutations,
metadata = sample_metadata,
response = MSI_status,
gbuild = 'BSgenome.Hsapiens.UCSC.hg38',
mut_sigs = 'COSMIC_v2_SBS_GRCh38',
tri.counts.method = 'default',
colors = c('orange', 'black'),
col.names = TRUE)GEGVIC offers the possibility to execute all the functions of a specific module at once using a single function with all the parameters described before. To access the result tables the output needs to be saved in a new object.
# Gene Expression Module (GE)
tables_module_ge <- module_ge(counts = sample_counts,
genes_id = 'ensembl_gene_id',
metadata = sample_metadata,
response = MSI_status,
design = 'MSI_status',
colors = c('orange', 'black'),
ref_level = c('MSI_status', 'MSS'),
shrink = 'apeglm',
biomart = ensembl_biomart_GRCh38_p13,
fold_change = 2,
p.adj = 0.05,
gmt = 'inst/extdata/c2.cp.reactome.v7.5.1.symbols.gmt',
gsea_pvalue = 0.2,
gsva_gmt = 'hallmark',
method = 'gsva',
kcdf = 'Gaussian',
row.names = TRUE,
col.names = TRUE)
# Genomic Variations Module (GV)
tables_module_gv <- module_gv(muts = sample_mutations,
metadata = sample_metadata,
response = MSI_status,
top_genes = 10,
specific_genes = NULL,
colors = c('orange' ,'black'),
compare = 'wilcox.test',
p_label = 'p.format',
gbuild = 'BSgenome.Hsapiens.UCSC.hg38',
mut_sigs = 'COSMIC_v2_SBS_GRCh38',
tri.counts.method = 'default',
col.names = TRUE)
# Immune cell Composition module (IC)
tables_module_ic <- module_ic(counts = sample_counts,
genes_id = 'ensembl_gene_id',
biomart = ensembl_biomart_GRCh38_p13,
indications = rep('coad', ncol(sample_counts[-1])),
cibersort = NULL,
metadata = sample_metadata,
response = MSI_status,
compare = 'wilcox.test',
p_label = 'p.format',
colors = c('orange', 'black'),
points = TRUE)Finally, GEGVIC offers the possibility to generate an HTML report which
will contain all the graphical outputs already shown in this manual
using the auto_rep() function. Appart from all the options already
explained for each function, the user has the option to select which
modules will be executed and also the output directory where the report
will be saved.
auto_rep(ge_module = TRUE,
gv_module = TRUE,
ic_module = TRUE,
out_dir = NULL,
counts = sample_counts,
genes_id = 'ensembl_gene_id',
metadata = sample_metadata,
response = 'MSI_status',
design = 'MSI_status',
colors = c('orange', 'black'),
ref_level = c('MSI_status', 'MSS'),
shrink = 'apeglm',
biomart = ensembl_biomart_GRCh38_p13,
fold_change = 2,
p.adj = 0.05,
gmt = 'c2.cp.reactome.v7.5.1.symbols.gmt',
gsea_pvalue = 0.2,
gsva_gmt = 'hallmark',
method = 'gsva',
kcdf = 'Gaussian',
row.names = TRUE,
col.names = TRUE,
muts = sample_mutations,
top_genes = 10,
specific_genes = NULL,
compare = 'wilcox.test',
p_label = 'p.format',
gbuild = 'BSgenome.Hsapiens.UCSC.hg38',
mut_sigs = 'COSMIC_v2_SBS_GRCh38',
tri.counts.method = 'default',
indications = rep('coad', ncol(sample_counts[-1])),
cibersort = NULL,
tumor = TRUE,
rmgenes = NULL,
scale_mrna = TRUE,
expected_cell_types = NULL,
points = TRUE)