Identification of disease treatment mechanisms through the multiscale interactome
Overview
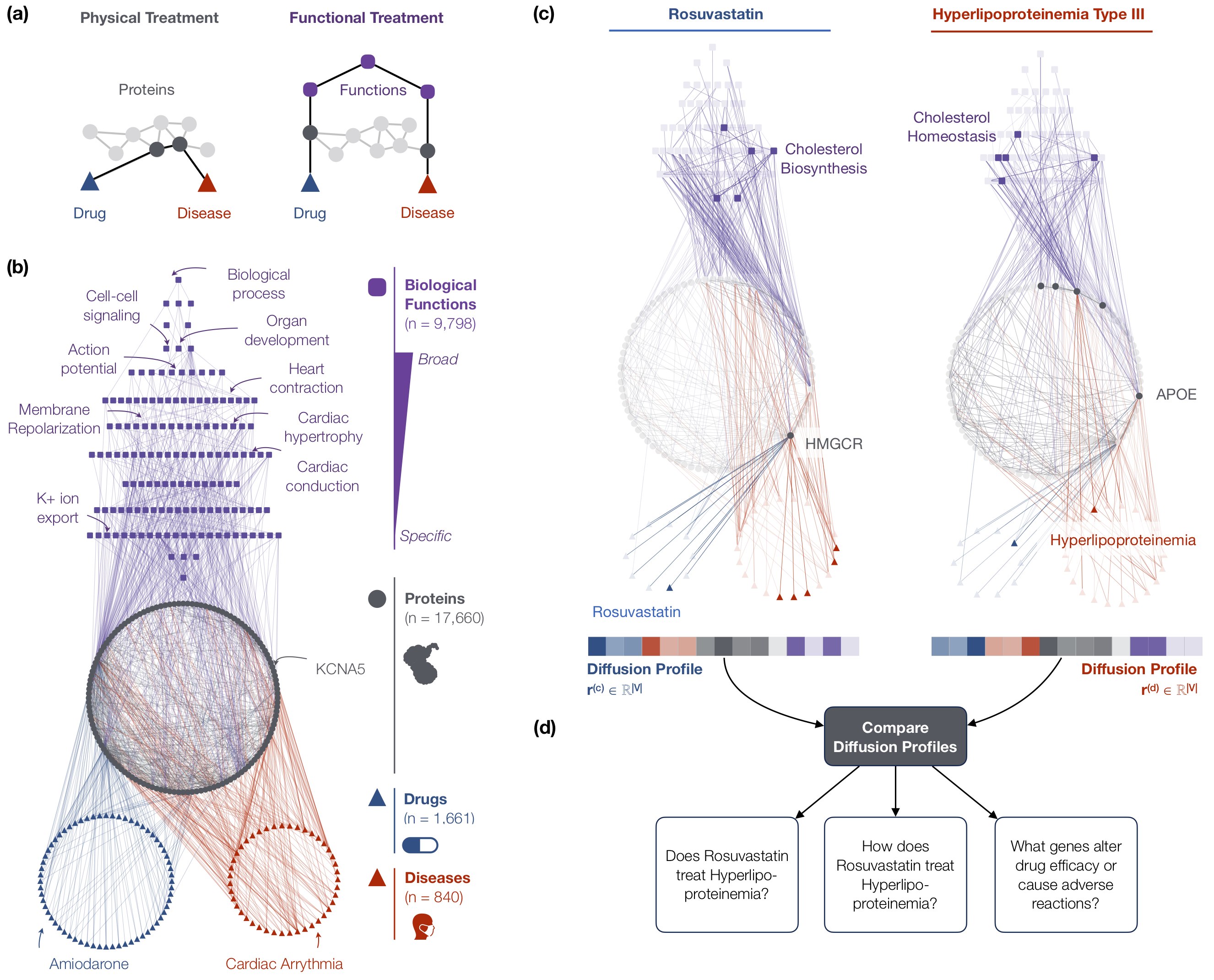
The multiscale interactome is a powerful approach to explain disease treatment. (a) Existing network approaches assume that drugs treat diseases by targeting proteins that are close to disease proteins in a network of physical interactions. However, drugs can also treat diseases by targeting distant proteins that affect the same biological functions. (b) The multiscale interactome models drug-disease treatments by integrating both proteins and a hierarchy of biological functions. (c) The diffusion profile of a drug or disease captures its effect on every protein and biological function. (d) By comparing the diffusion profiles of a drug and a disease, the multiscale interactome predicts drug-disease treatment, identifies proteins and biological functions related to treatment, and predicts genes that alter a treatment's efficacy and adverse reactions.
For a detailed description of the multiscale interactome and its applications, please refer to our preprint Identification of disease treatment mechanisms through the multiscale interactome (2020).
Data
All data is available at http://snap.stanford.edu/multiscale-interactome/data/data.tar.gz. To download the data, please run the following code in the same directory this project is cloned to. This should result in a data/ folder populated with the relevant data.
wget http://snap.stanford.edu/multiscale-interactome/data/data.tar.gz
tar -xvf data.tar.gz
Setup
Code is written in Python3. Please install the packages present in the requirements.txt file. You may use:
pip install -r requirements.txt
Using the multiscale interactome
Sample code to construct the multiscale interactome and compute diffusion profiles is provided in main.ipynb.
To construct the multiscale interactome:
msi = MSI()
msi.load()
To calculate diffusion profiles with optimized edge weights, use the code below. Note that our implementation is parallelized and you will need to explicitly set the number of cores under num_cores.
dp = DiffusionProfiles(alpha = 0.8595436247434408, max_iter = 1000, tol = 1e-06, weights = {'down_biological_function': 4.4863053901688685, 'indication': 3.541889556309463, 'biological_function': 6.583155399238509, 'up_biological_function': 2.09685000906964, 'protein': 4.396695660380823, 'drug': 3.2071696595616364}, num_cores = int(multiprocessing.cpu_count()/2) - 4, save_load_file_path = "results/")
dp.calculate_diffusion_profiles(msi)
To load saved diffusion profiles, use:
dp_saved = DiffusionProfiles(alpha = None, max_iter = None, tol = None, weights = None, num_cores = None, save_load_file_path = "results/")
msi.load_saved_node_idx_mapping_and_nodelist(dp_saved.save_load_file_path)
dp_saved.load_diffusion_profiles(msi.drugs_in_graph + msi.indications_in_graph)
To view a diffusion profile, use:
# Diffusion profile for Rosuvastatin (DB01098)
dp_saved.drug_or_indication2diffusion_profile["DB01098"]
Datasets
All data is provided at http://snap.stanford.edu/multiscale-interactome/data/data.tar.gz and includes:
- Supplementary dataset of interactions between drugs and proteins
- Supplementary dataset of interactions between diseases and proteins
- Supplementary dataset of interactions between proteins and proteins
- Supplementary dataset of interactions between proteins and biological functions
- Supplementary dataset of hierarchy of interactions between biological functions
- Supplementary dataset of approved drug-disease pairs
- Supplementary dataset of drug classes according to Anatomical Therapeutic Chemical Classification
- Supplementary dataset of selected gene expression signatures from the Broad Connectivity Map
- Supplementary dataset of genes that alter drug response from PharmGKB
- Supplementary dataset of optimized diffusion profiles
Contact
Please contact Camilo Ruiz (caruiz@cs.stanford.edu) with any questions.
Citation
@article{ruiz2020identification,
title={Identification of disease treatment mechanisms through the multiscale interactome},
author={Ruiz, Camilo and Zitnik, Marinka and Leskovec, Jure},
journal={bioRxiv},
year={2020},
publisher={Cold Spring Harbor Laboratory}
}