https://appliedmldays.org/schedule
The repos for the tutorials I attended plus a few extras can be found here. Some other tutorial repos are stored here: https://github.com/amld.
Other random notes below. See also Some NLP references.
Some other interesting conferences:
https://www.eyeforpharma.com/barcelona/conference-agenda.php
https://www.oxfordglobal.co.uk/pharmatec-series/
-
- 2.1. Interpretability
- 2.1.1. Gradient-based methods
- 2.1.2. Perturbation-based methods
- 2.1.3. Adversial robustness
- 2.1.4. Rule-based models
- 2.1.5. Example-based
- 2.1.6. Shapely
- 2.1.7. Surrogate methods
- 2.1.8. Multi-interpretation problems
- 2.2. Libraries
- 2.1. Interpretability
-
- 3.1. Torch Tensors
- 3.2. Autograd
- 3.3. Optimisation
- 3.4. CNNs
-
- 4.1. Intro
- 4.2. Using BERT
- 4.2.1. Several methods are possible
- 4.2.2. Issues with BERT
- 4.2.3. Excerise 1 - Finetuning BERT IMDB sentiment analysis
- 4.2.4. Tokenization, Word Pieces and out-of-vocabulary words
- 4.2.5. Padding
- 4.2.6. Task
- 4.2.7. Results
- 4.3. Transfer Learning At Scale
- 4.3.1. Tokenization
- 4.3.2. Distillation
- 4.4. Quantization
- 4.5. Serving models at scale
- 4.6. Correlation vs Causation
-
- 6.1. ML without growing team at Swisscom
- 6.2. Dataiku
- 6.3. Learning Actionable Representations of Biomedical Data, Marinka Zitnik
- 6.3.1. Example - drug repurposing
- 6.4. Roche: Making Personalised Healthcare a reality? – More than AI, Asif Jan
- 6.5. Buhler - Data Science for sustainable manufacturing
- 6.6. Swiss, Re Institute. Bohn: Addressing the failure of enterprise machine intelligence (MI) with 4 AIs we should discuss more
- 6.7. Unity: Simulations – The New Reality for AI, Danny Lange
- 6.8. Microsoft and Novartis: Machine Learning at Exa-scale: Opportunities for Life Sciences, Shahram Ebadollahi & Christopher Bishop
-
- 7.1. Education - Enhancing human learning via spaced repetition optimization, Manuel Gomez Rodriguez
- 7.2. Health - Roche - Mapping medical vocabulary terms with word embeddings, Pekka Tiikkainen
- 7.3. Imaging - EMBL-EBI - Machine Learning for Bioimage Informatics: Successes, Challenges, and Perspectives, Virginie Uhlmann
- 7.3.1. Image-based modelling
- 7.4. Health - Beyond lesion count: machine learning based imaging markers in multiple sclerosis
-
- 8.1. Vetterli
- 8.2. Kosinski - The End of Privacy
- 8.3. Tegmark
-
- 10.1. Roche Intro - Asif Jan
- 10.2. Developing Digital Measures from Person-Generated Health Data, Luca Foschini, Co-founder & Chief Data Scientist, Evidation Health
- 10.3. Retinai - Detection and quantification of disease biomarkers in ophthalmology, Agata Mosinska
- 10.4. Astra Zeneca Bayesian Neural Networks for toxicity prediction, Elizaveta Semenova
- 10.5. Roche - A Machine Learning perspective on the emotional content of Parkinsonian speech, Konstantinos Sechidis
- 10.6. U. Manchester - On the Stability and Reproducibility of Data Science Pipelines, Gavin Brown
- 10.7. Turing Institute - Going beyond the average: causal machine learning for treatment effect heterogeneity estimation, Karla DiazOrdaz
- 10.8. Novartis - AE Brain - Detecting Adverse Drug Events with NLP, Damir Bucar & Moritz Freidank.
- 10.9. Novartis - Fully unsupervised deep mode of action learning for phenotyping high-content cellular
- 10.10. Novartis - Benchmarking initiative: making sense of AI through the common task framework, Mark Bailli
- 10.11. Roche - Artificial Intelligence in clinical development. Marco Prunotto
-
- 11.1. BAIR: Jenny Listgarten - Machine Learning-based Design of Proteins and Small Molecules
- 11.2. U Geneva: Conditional Generation of Molecules from Disentangled Representations. Amina Mollaysa
- 11.3. LBNL An Autoencoder for 3D Geometries of Atomic Structures with Euclidean Neural Networks, Tess E. Smidt
- 11.4. EPFL: Physics-inspired Machine Learning for Materials Discovery, Michele Ceriotti
- 11.5. Google DeepMind: AlphaFold - Improved protein structure prediction using potentials from deep learning, Andrew Senior
- 11.5.1. Protein Co-evolution
- 11.5.2. Deep distance distribution network
- 11.6. U. Liverpool Data Driven Discovery of Functional Molecular Co-Crystals, Aikaterini Vriza.
- 11.7. LabGenius - Knitting together synthetic biology, machine learning and robotics, Katya Putintseva
- 11.7.1. Protein Fitness Landscape
- 11.7.2. Main challenge
-
- 12.1. Anomaly detection with Isolation Forests
- 12.2. Text classification
- 12.3. Image Classification
- 12.4. Facial recognition in videos
-
Saturday AM: Interpretability in Comp Bio <https://github.com/IBM/depiction/tree/master/workshops/2020012 _AMLD2020>
-
Saturday PM: PyTorch https://github.com/theevann/amld-pytorch-workshop
-
Sunday AM: Transfer Learning at scale in Natural Language Processing
-
Sunday PM: Cross-Lingual NLP terrible code, interesting material
-
Monday AM: Keynotes
-
Monday PM:
-
Tuesday AM: 4BC https://appliedmldays.org/tracks/ai-pharma
-
Tuesday PM: 1A https://appliedmldays.org/tracks/ai-molecular-world
- Note there's also
- NLP https://appliedmldays.org/tracks/ai-nlp
- Zitnik, Large-scale prediction of phenotypes from biological networks 1 -:30 - 16:10, 2A, AI & Nutrition
- Note there's also
-
Wednesday AM: Keynotes https://appliedmldays.org/tracks/wednesday-keynote-session
-
Wednesday PM: ??
- Governance 2 panels https://appliedmldays.org/tracks/ai-governance
- Skills https://appliedmldays.org/tracks/ai-skills
Ancona et al, Towards better understanding of gradient-based attribution methods for Deep Neural Networks
https://arxiv.org/abs/1711.06104
Marco Tulio Ribeiro, Sameer Singh, Carlos Guestrin, "Why Should I Trust You?": Explaining the Predictions of Any Classifier
https://arxiv.org/abs/1602.04938
Ribiero, Anchors:
https://homes.cs.washington.edu/~marcotcr/aaai18.pdf
Attention as interpretabilty: interpret alpha's like beta in linear model. See Bengio et al: "learning to align and...""
Girardi,Patient Risk Assessment and Warning Symptom Detection Using Deep Attention-Based Neural Networks
https://arxiv.org/abs/1809.10804
Explanations based on the Missing: Towards Contrastive Explanations with Pertinent Negatives, Amit Dhurandhar, Pin-Yu Chen, Ronny Luss, Chun-Chen Tu, Paishun Ting, Karthikeyan Shanmugam, Payel Das
https://arxiv.org/abs/1802.07623e
Counterfactual Explanations without Opening the Black Box: Automated Decisions and the GDPR, Sandra Wachter, Brent Mittelstadt, Chris Russell
https://arxiv.org/abs/1711.00399
Generalized Linear Rule Models DennisWei1 Sanjeeb Dash TianGao Oktay Gunluk http://proceedings.mlr.press/v97/wei19a/wei19a.pdf
"This looks like that"
Calculates payout player/feature deserves
IBM upcoming article on modelling how to combine expert interpretation/annotation.
Depiction
https://kipoi.org - omics model view.
https://science.sciencemag.org/content/332/6030/687/tab-figures-data
https://github.com/theevann/amld-pytorch-workshop
torch.tensor behaves like numpy arrays under mathematical operations. However, torch.tensor additionally keeps track of the gradients (see next notebook) and provides GPU support.
CAUTION: Operator * does element-wise multiplication, just like in numpy!
Y.t() * Y # error, dimensions do not match for element-wise multiplication. As does + etc.
Each PyTorch tensor operation is also available as a function.
Functions that mutate (in-place) the passed object end with an underscore, e.g. add_, div_, etc.
Indexing works like in numpy.
view(*shape) → Tensor
Returns a new tensor with the same data as the self tensor but of a different shape. The returned tensor shares the same data and must have the same number of elements, but may have a different size. -1 tells PyTorch to infer the number of elements along that dimension. Equivalent of 'reshape' in numpy (view does not allocate new memory!)
expand() # similar to repeat but does not actually allocate new memory
squeeze() # removes all dimensions of size '1'
.backward() accumulates the gradients. If you want fresh values, you need to set x.grad to zero before you call .backward().
In the example below optimizer keeps track of what you need:
0 = 8
lr = 0.1
iterations = 10
x = torch.Tensor([x0]).requires_grad_()
y = f(x)
# Define your optimizer
optimizer = optim.Adam([x], lr=lr)
for i in range(iterations):
y = f(x)
y.backward() # compute gradient
optimizer.step() # does everything - knows the function x
optimizer.zero_grad()
print(y.data)Note that in nn architectures, the softmax activation function of the final hiiden layer goes in the loss function in Pytorch convention:
class CrossEntropyLoss(_WeightedLoss):
r"""This criterion combines :func:`nn.LogSoftmax` and :func:`nn.NLLLoss` in one single class...First kernel gives vertical lines. Transpose to get horizontal lines.
Need as many kernels as output channels.
HuggingFace Transformers library being used.
Bag of words: simple count, sparse representation.
Embeddings: dense representations.
Word2vec: Distributional Hypothesis (1954): “a word is characterized by the company it keeps” Static embedding methods are not very good at encoding the context. One vector for one word. RNN weight sharing
RNN: information needs kept for a long time in internal states.
Attention: importance of every token for every other token - Query Key product (care: matrix vis in slides is random!)
BERT introduced bi-directionality by using a transformer instead of an LSTM encoder
Uses two objectives: Mask Language Model (MLM) next sentence prediction (NSP)
Uses special tokens to format the input: [CLS], [SEP], [MASK]
Frozen BERT + task layers
Adapters: “Parameter-efficient Transfer Learning for NLP” http://proceedings.mlr.press/v97/houlsby19a.html Adapters ⇒ obtain good accuracy with less weights to tune! Catastrophic forgetting: forgetting what you learn in the pre-training task. Use small learning rates to avoid big changes or regularization. ULMfit: Especially for lower layers which capture general information (https://arxiv.org/abs/1801.06146). Learning rate warmup.
-
Freezing everything but the top layer (https://arxiv.org/abs/1502.02791)
-
Training one layer at a time (https://www.aclweb.org/anthology/D17-1169/)
-
Gradually unfreezing (https://arxiv.org/abs/1801.06146)
-
Fine-tune all parameters.
What/when to choose: “To Tune or Not to Tune? Adapting Pre-trained Representations to Diverse Tasks”, https://arxiv.org/pdf/1903.05987.pdf
The feedforward pass is slow (Bert Large has 335M parameters)
Fine-tuning is very unstable (https://arxiv.org/pdf/1811.01088.pdf)
Memory problems if the maximum sequence length is long
You can pre-train your own version of Bert on your dataset Requires lots of GPUS/TPUS Tricky to choose hyper-parameters
Tested a bit on mac. Moved over to colab for GPU:
Here are some of the main parameters you will want to consider when fine-tuning BERT:
-
Gradient Clipping: If the norm of the gradient gets above max_grad_norm, We divide the gradient by its L2 norm. This gradient clipping method avoids exploding gradients.
-
Learning rate: The learning_rate parameter is very important as it controls how we update the already trained parameters from the Language Modelling Task. If this parameter is too high, we will notice a forgetting of the previous task. It needs to be carefully tuned.
-
Sequence length: The attention mechanism scales in O(L^2). So you should avoid handling sequences larger than what you really need.
BERT uses Word pieces. Tokens are sequences of characters - optimized. This is useful for out of vocabulary words. Word pieces are kinda of doing stemming.
The input to the BERT model are word-pieces (Original paper). Standard tokens are broken down into word pieces through the use of a WordPiece tokenizer.
A WordPiece tokenizer breaks the unknown words into multiple subwords. For example, if the word "chore" does not belong to the vocabulary as a single piece, it might get split into two pieces belonging to the vocabulary: 'cho' and '##re'.
All the subwords start with the "#" symbol except for the first subword in the word. Imagine the words "played", "playing" are rare words and thus would not occur in a normal vocabulary. These words would be considered into the wordpiece tokenizer into this form: [play, ##ed] and [play, ##ing].
You can have a look at the file bert-base-uncased-vocab.txt in your environment to have an idea of the words present in the vocabulary
Wordpiece tokenizers tends to be quite slow, however some efficient implementations exist: tokenizers from the Huggingface Library are much faster than the standard naive implementations (Implemented in Rust with python bindings).
There is padding to max sequence length
Note [CLS] token is always appearing first for the classification task.
768 - is the bert hidden layer size
Note finetuning loop is very simple: no early stopping etc.
Spacy tokenizer is the simple classifier user. BERT is much better for F1-score, but not other metrics covering speed and size etc.
HuggingFace Tokenizer is much faster.
Same data point goes to student and teacher. Teacher not trained, only student. Student training tries to mimic student (student loss).
Approach 1: Replace label with Teacher predictions. Allows us to apply to much more unlabeled data
Approach 2: Uses labels. MSE on logits plus cross-entropy loss. Some work “Distilling Task-Specific Knowledge from BERT into Simple Neural Networks”: https://arxiv.org/abs/1903.12136v1 showed that the best performance had the cross-entropy turned off!
Why quantize:
-
Models benefit from hardware optimizations → faster
-
Significantly reduce the memory footprint! → Smartphone friendly!
-
On deep network, quantization usually do not significantly degrade the performances
-
BERT and other large model inference on CPU is very slow. Quantized version is faster.
See: “quantization-in-tflite” https://sahnimanas.github.io/post/quantization-in-tflite/
and: “Pytorch quantization” https://pytorch.org/docs/stable/quantization.html
Models can be quantized post-training and during training.
-
During training: Quantize weights in the forward pass Backward pass and the update are with regard to the unquantized weights
-
Post-training: Use unquantized weights while training Quantize weights after training and perform inference
Quantization is very simple:
import torch.quantization
def print_size_of_model(model):
"""""
Get the size on disk of the model
"""""
torch.save(model.state_dict(), "temp.p")
print('Size (MB):', os.path.getsize("temp.p")/1e6)
os.remove('temp.p')
bert.eval()
bert.cpu()
# As simple as that:
quantized_bert = torch.quantization.quantize_dynamic(
bert, {nn.Linear}, dtype=torch.qint8
)Quantized BERT took 61 seconds whereas full BERT took 78 seconds.
Note the scores aren't great in the notebook just because we took only 100 datapoints to save time.
-Containerize your model to make replication easy
-Use spot instances (Cost savings)
-Orchestrate your containers for autoscaling
-Handle the serving, logging, monitoring, ...
One open source solution on AWS:
Clever Hans - attending to specifc signals but not learning. Entailment.
https://thegradient.pub/nlps-clever-hans-moment-has-arrived/
“Right for the Wrong Reasons: Diagnosing Syntactic Heuristics in Natural Language Inference” https://www.aclweb.org/anthology/P19-1334.pdf
-
The paper shows that Bert “closely tracks statistical regularities in the training set” → Which is prone to adopting superficial heuristics
-
Good test data is the most valuable there is! → It’s ok to have a test and train dataset from different distribution if the test data is better matching what you’re aiming for.
-
A lot of work need to be done to understand what models are exactly learning
Catastrophic code failures! I've deleted any local work. Just go and try to make sense out of the files on my github.
https://github.com/ioannispartalas/CrossLingual-NLP-AMLD2020
Good for low resource langauges.
Vectorization: semantically similar words can have different vectors.
Text representation: traditional bag-of-words. Given a text, extract the vocabulary, build a vector of dim
https://arxiv.org/abs/1706.04902
https://arxiv.org/abs/1812.10464
Ethics: 'original why it was possible' instead of regulatory. Policy people also need to know ML.
https://www.heidi.news/newsletters
https://www.remotecon.org/index.html
https://www.extensionschool.ch/
sli.do AMLD2020 https://app.sli.do/event/uldojaf3/live/questions
What comes after breaking the silos (last year). So many ops. How to prioritise?
-
What else is this data good for?
-
How can integrate constraints and objectives? Fairness.
-
I am using everything relevant?
Multiobjective sgd requires gradient normalization.
Evaluating the Search Phase of Neural Architecture Search Kaicheng Yu et al: https://arxiv.org/abs/1902.08142
Pitch for product
https://stanford.edu/~marinka/
videolectures.net › icgeb_zitnik_graph_neural_networks Graph neural networks for computational drug repurposing
Overview of ops for ML in health and medicine, but it's challenging:
-
Data integration, confounded, scales
-
Actionable outputs.
Networks and data integration: Population - individuals - molecular. AD mentioned
Actionable outputs: target id, treatment.
Bipartite graph of drug-disease interactions.
Disease as subgraph of rich protein network on disease proteins.
Drug as subgraph of rich protein network on drug's trarget proteins.
Drug likely to treat disease if close in pharamcological space, but what does this mean.
Method: SUGAR.
Graph NN, neural message passing.
Stanford SPARK translational gave drug disease repurposing data - supervised.
Highlight what part of molecular structure are important
GNNExplainer: Generating Explanations for Graph Neural Networks
Neurips: https://papers.nips.cc/paper/9123-gnnexplainer-generating-explanations-for-graph-neural-networks.pdf
To Embed or Not: Network Embedding as a Paradigm in Computational Biology https://www.frontiersin.org/articles/10.3389/fgene.2019.00381/full
Deep Learning for Network Biology: http://snap.stanford.edu/deepnetbio-ismb/
https://www.iscb.org/ismb2020-submit/proceedings
Asking the right q, getting the right data. Collaborate.
Electronic medical records. RWE.
EDIS data mart. Clinical, omics and imaging. >40k patients. FAIR data, Wilkinson ete al Scientific Data 2016:3.
Flatiron partner. Foundation medicine.
Carrigan et al, Clin Pharmacol Ther 2019.
Using RWE as control data to reduce burden on control arm: Using Electronic Health Records to Derive Control Arms for Early Phase Single-Arm Lung Cancer Trials: Proof-of-Concept in Randomized Controlled Trials. https://www.ncbi.nlm.nih.gov/pubmed/31350853
PicnicHealth MS. Williams et al ECTRIMS 2019.
Curtis et al Health Surv Res 2018: 53.
Improved curation and quality (manual and auto) to improve results.
RANN: Roche advanced analytics data network. Crowd sourced across Roche. Win lunch with CEO. Lots of good models - ensembles.
Digital twin. Replay productions. Mobile rice quality monitor.
6.6. Swiss, Re Institute. Bohn: Addressing the failure of enterprise machine intelligence (MI) with 4 AIs we should discuss more
Lots of failures. Bad planning. A load of guff. Gartner.
binderhub
Need to explain in 'ai'
- hyper focussed.
- integrate in business
- not regular it projects
- mlops
- right tool for q
- hr
4ais:
-
Augmented
- human in the loop
-
Automation
- data ingestion and curation
- ml for upstream data clearning etc.
- mlops
- can lead to glaring mistakes. human-in-loop
-
Assessed Intelligence
- prefer robustness, stability, interpretability
-
Adaptive Intelligence
- physical models, RL
- behaviour mimicking?
Causal inference.
Simulations for annotation and dataset creation. 100% true ground truth? But who fixed the physics/simulations and what may be missed.
RL: explore v exploit. observe, action, reward.
RL robots. Peter Pastor. Too long. Need simulated data.
Planck time?!
Beating evolution
- faster than 30 fps (human preferred)
- parallelism
- blend species and individual
- step back in time (checkpointing)
- lower frequency: drop fill frames. But who decides?
6.8. Microsoft and Novartis: Machine Learning at Exa-scale: Opportunities for Life Sciences, Shahram Ebadollahi & Christopher Bishop
3 key elements
- Data
- No free lunch. Prior knowledge. Less data, more prior knowledge needed.
- Massive compute.
The Bitter Lesson - Sutton.
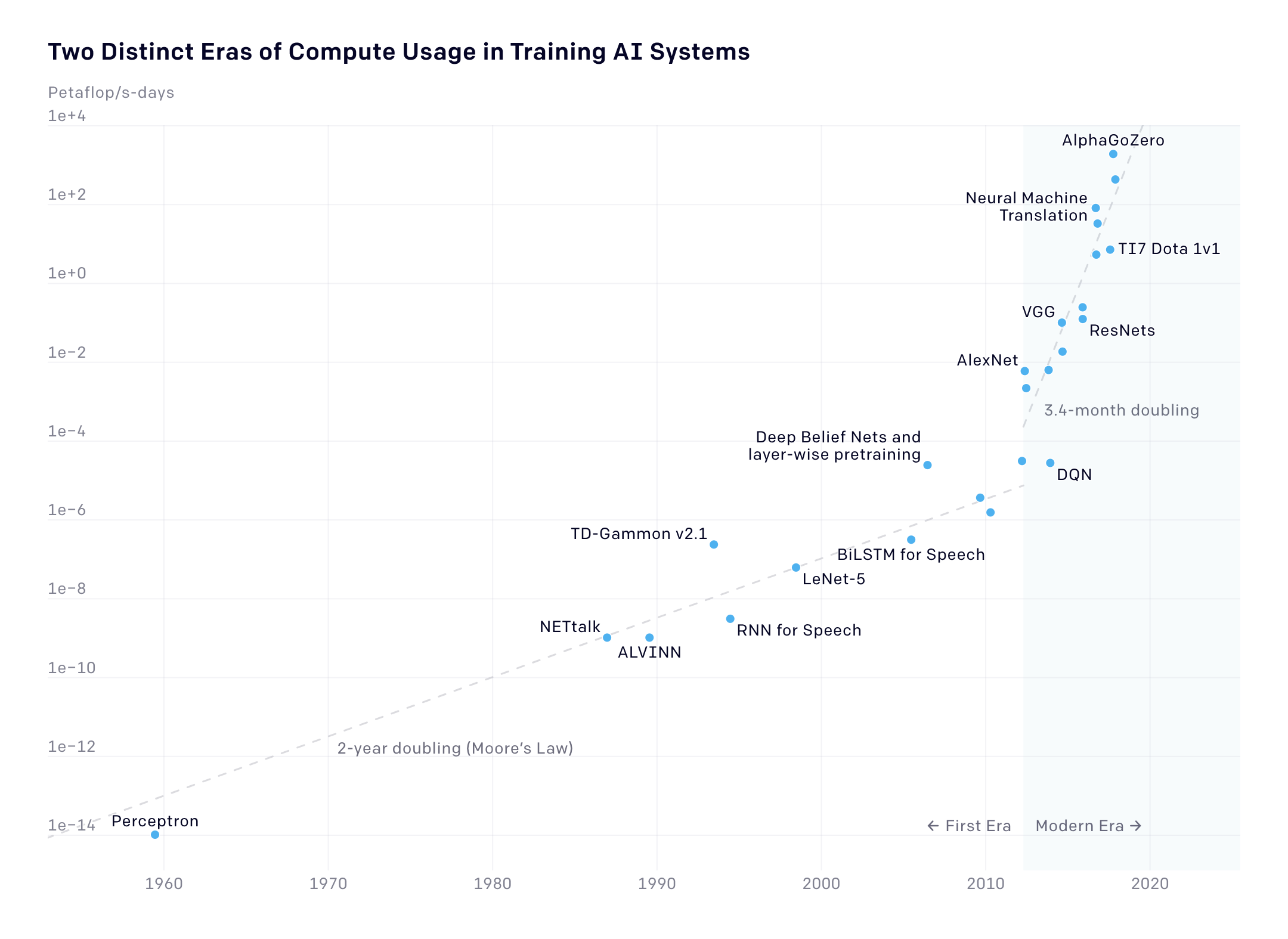
OpenAI compute over time graph https://openai.com/blog/ai-and-compute/:
Exponential growth up to 2010 in ML SOTA compute (Moores law of transistors -> compute). Since 2010, doubling time dropped to 3.5 months. 10x increase every 12 months.
5mb - complete works of shakespear. cf BERT.
Imperative to stay on trajectory. Huge ask, few companies. Microsoft invested in OpenAI, who move to MS for exa-scale compute. How? e.g Graphcore 23.6 billion transistors.
Cholera example. John Snow. Epidemiology invented.
Novartis
https://dblp.org/pers/hd/e/Ebadollahi:Shahram
Body as complex data generator.
Data42. 2x10ˆ6 patient year of data. Lots about disease progression and longitudinal studies.
Challenges:
-
Drug design. Space of all chem compounds 10ˆ60. Still heavily expert driven. HT assays and compounds. Graph NN for molecule. Message passing. NeurIPS 18. Autoencoder for molecule. How smooth is this space. Framed as Netlix recommender with parameters fixed.
-
Age related macular disease. Vision loss. CV augmented human vision. Deep CV on eye images for precision timing and dosing of drug.
400 people hackathon for P(success). 12 month fellowships for ML non-bio people.
https://dblp.org/pers/hd/e/Ebadollahi:Shahram
7.1. Education - Enhancing human learning via spaced repetition optimization, Manuel Gomez Rodriguez
https://https://ari9000.com/ code: AMLD
Leitner systems for cards.
Modelling decay of memory. Half-life regression. It looks like this could use survival analysis. Censoring?
Stochastic optimal control for review.
Drug safety department. e.g. what is baseline event rate? ICD9-CM + 10. MedDRA RWD databases. Need to map vocabulary, often manually done before.
Use word embeddings. word2vec CBOW (context-> word) or skip-gram (word -> context).
Used MIMIC3 and Roche internal data.
Baseline: term concurence. See photo.
Pancreatic cancer often metstasticise in lungs, hence appearance in lung results.
fasttext and word2vec outperformed baseline. Corpus choice didn't change much.
7.3. Imaging - EMBL-EBI - Machine Learning for Bioimage Informatics: Successes, Challenges, and Perspectives, Virginie Uhlmann
Prepared with Anne Carpenter, Jan Funke, Florian Jub, Anna Krushuk.
Huge datasets being generated. Each image is information dense. Keller lab: <Digital-embryo.org>. Cryo-EM Toro-Nahuelpan et al, Nature 2019, Tailoring cryo-electron microscopy grids by photo-micropatterning for in-cell structural studies, https://www.biorxiv.org/content/10.1101/676189v1
Want: quantitative, scalable and reproducible.
Bioimaging - each experiment has its own kind of images. Question -> find something!
ilastik - rf classifier looks very powerful. Kreshuk group.
Image restoration, image preprocessing, image based modelling.
Martin Weigert et al. Nature Methods 2019, Content-aware image restoration: pushing the limits of fluorescence microscopy, https://www.nature.com/articles/s41592-018-0216-7 Use high-light low noise regions to learn how to make high noise, low light. Then reverse.
Noise2void: Krull et al CVPR 19. Learn what is correlated (signal) and what is uncorrelated (noise), then remove noise.
Funke lab - Image understanding. EM images. Neuron tracing.
Buhmann et al, Automatic Detection of Synaptic Partners in a Whole-Brain Drosophila EM Dataset, https://www.biorxiv.org/content/10.1101/2019.12.12.874172v1. Eckstein et al. Neurotranslitter classification. Predict what kind of neurotransmitter is released in some regions.
Kruskuk et al: Complete Cryo-EM imaging of platynereis flat worm. 8TBs. 12k cells.
Soham Mandal et al, biorxiv 19. CNN- > morphology. Diff. geometry. eg 6-cell stage mouse embryo. Hiiragi group EMBL. Deformation infers cell function? Diffeomorphic mapping? Sharpe group, Barcelona. .
Carpenter lab: Cell Profiler. Images for drug discovery. Juan. Caicedo et al. CVPR 18, Weakly Supervised Learning of Single-Cell Feature Embeddings, http://openaccess.thecvf.com/content_cvpr_2018/html/Caicedo_Weakly_Supervised_Learning_CVPR_2018_paper.html What is the latent representation learnt? Note that annotators didn't know that mutations causing cancer. Network manages to identify mutation clusters.
See also Rohban et al, mental health work. Distribution in low dim space is different depending on norma and disorders.
Challenges: Annotation, integrate omics, correlate imaging techniques.
Community : image.schttps://forum.image.sc/
Nature Methods: Deep Learning in Microscopy.
Segmentation still a big issue.
Liu et al Lancet Digital Health, A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. https://www.thelancet.com/journals/landig/article/PIIS2589-7500(19)30123-2/fulltext
Most MS lesions have a vein in the middle - how the disease propagates.
Ravano et al disconectome ECTRIMS 19.
Ball: How to grow a human.
RUR Kapek
500mb digital footprint everyday in 2012 (IBM).
3 traits link personality and face.
Hormones affect faces.
Traits from faces: People get about 54% in lab test. Think about how gender can be determined. Brain just not evolved to this yet. Computer hits work colleague level of accuracy.
Predicting political views from faces: 90% accurate.
Privacy is already lot. We need to make the post-privacy world livable. The problem is not privacy, it is e.g. homophobia.
Ultrasound for everyone: https://www.butterflynetwork.com/
Asilomar 2017, Principles: https://futureoflife.org/ai-principles/?cn-reloaded=1
AI Feynman: a Physics-Inspired Method for Symbolic Regression Silviu-Marian Udrescu (MIT), Max Tegmark (MIT) (Submitted on 27 May 2019) A core challenge for both physics and artificial intellicence (AI) is symbolic regression: finding a symbolic expression that matches data from an unknown function. Although this problem is likely to be NP-hard in principle, functions of practical interest often exhibit symmetries, separability, compositionality and other simplifying properties. In this spirit, we develop a recursive multidimensional symbolic regression algorithm that combines neural network fitting with a suite of physics-inspired techniques. We apply it to 100 equations from the Feynman Lectures on Physics, and it discovers all of them, while previous publicly available software cracks only 71; for a more difficult test set, we improve the state of the art success rate from 15% to 90%.
https://arxiv.org/abs/1905.11481
https://rodneybrooks.com/blog/
AI in sustainable development goals. Tegmark et al.
Pytorch lightening preferred high level abstraction by NLP at scale guys. See also Pytorch ignite.
https://github.com/flairNLP/flair
https://ml6.eu/a-general-pattern-for-deploying-embedding-based-machine-learning-models/
General overview.
10.2. Developing Digital Measures from Person-Generated Health Data, Luca Foschini, Co-founder & Chief Data Scientist, Evidation Health
Person Generated Health Data. Toward achieving precision health. Gambhir SS, https://www.ncbi.nlm.nih.gov/pubmed/29491186. ALso includes patient reported data.
path collaborative wearable tech landscape.
What happens in between visits to the doctor.
Evidation Achievement - anyone can use with open data sharing. Can join clinical studies. Reconsent for every retro analysis for complete clarity.
Combination of passive and task-based data collected.
Behaviourgram - find a way to actually look at your data! Inspect data quality and generated hypotheses to test. Most useful part of study.
Can you differentiate cohorts based on behaviourgram. Features engineered in usual way. xgboost used for predictions. Behaviorome.
SHAP used for interpretation.
https://evidation.com/research/
Structured data DL: https://towardsdatascience.com/structured-deep-learning-b8ca4138b848 etc.
Drug induced liver injury.
Preclinical models only transfer to humans in 45%: Concordance of toxicity... https://www.ncbi.nlm.nih.gov/pubmed/11029269
Deep-learning for drug-induced liver injury, Xu et al https://pubs.acs.org/doi/abs/10.1021/acs.jcim.5b00238
Interpretable Outcome Prediction with Sparse Bayesian Neural Networks in Intensive Care, Hiske Overweg. https://arxiv.org/abs/1905.02599
Moving beyond Binary Predictions of Human Drug-Induced Liver Injury (DILI) toward Contrasting Relative Risk Potential, Aleo MD https://www.ncbi.nlm.nih.gov/pubmed/31532188
Ordered logistic regression (increasing severity score).
Predicting Drug-Induced Liver Injury with Bayesian Machine Learning, Williams DP, https://www.ncbi.nlm.nih.gov/pubmed/31535850
Weights and biases described by distribution. Variances are being inferred from data.
Evaluation - WAIC. Ordered Brier Score - cares about the order.
150 parameters. Posterior calculated: prior, ordered likelihood (contributed this).
10.5. Roche - A Machine Learning perspective on the emotional content of Parkinsonian speech, Konstantinos Sechidis
PD perceived as being unhappier compared to control, negative impact to sufferer - isolation etc.
Combine mobile sensor data, treatment, speech etc. Annotated MP3 data. There are a few emotional speech databases. Often actors.
Standard feature engineering - 132 features. Catboost 2018. Mixture-of-Experts ensemble (k models).
train: gbm, val: lr, num trees, tree depth. stratified cross-val (of course!).
Weird choice to note combine databases 'to avoid biases' - I would have precisely combined the data for this reason. Maybe I'm missing something?
With Roche and AZ.
Thesis: Reproducibility=Trust. We can measure Repro.
Data-driven biomarker detection.
Drop percentages of data to measure stability. Set theoretic as its on features. But then he talks about dropping data points, which is features.
5 properties.
On The Stability of Feature Selection in the Presence of Feature Correlations: http://www.cs.man.ac.uk/~gbrown/publications/ECML2019_stability.pdf
10.7. Turing Institute - Going beyond the average: causal machine learning for treatment effect heterogeneity estimation, Karla DiazOrdaz
Causal effects of cancer treatments. Potential outcomes. Causal estimand. Statistical estimand from observed data. average treatment effect (ATE), conditional average treatment effect (CATE). https://arxiv.org/pdf/1903.00402.pdf
Intervals: bootstrap not good. Plug-in bias in ML. Doubly-robust estimators. See also targeted MLE.
Causal forest - https://scholar.princeton.edu/sites/default/files/bstewart/files/lundberg_methods_tutorial_reading_group_version.pdf
Maximise heterogeneity in treatment effect in terminal node, rather than rmse. "Honesty".
Double Machine Learning?
Monitoring social media text for potential cases of adverse events is an important, but manual and expensive task in pharmacovigilance. AE Brain uses an ensemble of NLP models (BERT, BioBERT, character-based models) to automatically detect potential mentions of adverse events and reduce the manual workload required for monitoring. By adopting a human-in-the-loop approach and rendering re-training fully reproducible our system ensures continuous improvement of our model and achieves GxP compliance.
Detect potential adverse events.
10ˆ5 posts per year, multiple sources.
GxP compliant but only with human-in-the-loop. Minimize FN rate.
Inter-annotator agreement as issue. Only gave results
Split tweet into sentence, then binary classifier. Forward to human reviewer. Class imbalance. Low annotator agreement. Implicit context. BERT and BioBERT. Bidirectionality clearly useful here.
512 max seq. length - very important, as high as you can get. batch size 4. 0.0001. 3 epochs. uncased. BioBERT worse overall, but good for edge cases. Ensembles used. Different random seeds for same model too. Any positive goes to human riew. All ensembled. Some rule based methods use for edge cases too.
Retraining for model drift. Use human review to feedback. Subsampled negatives too to feedback into retrain (ie those not seen by human).
English for now. Pointing
Pan-industry - looking for collaborators
10.9. Novartis - Fully unsupervised deep mode of action learning for phenotyping high-content cellular
images, Rens Janssens
No superivison on mode of action. Use unsupervised DL to look for clusters of mode of action. Multiscale CNN https://www.ncbi.nlm.nih.gov/pubmed/28203779 and Facebook AI clustering.
Multiscale has big field of few - local and overall cell population.
Remove last layer to get embeddings. Batch correaction, whitened, tse and clustered. DeepCluster https://arxiv.org/abs/1807.05520.
Tiled images
Clustered. Some separation is clear before training. Look at clusters epoch by epoch.
Leave out one mode of action and test. Good results.
10.10. Novartis - Benchmarking initiative: making sense of AI through the common task framework, Mark Bailli
Care with confoudning issues, e.g. picking up prior surgery marks.
Large landscape: https://techburst.io/ai-in-healthcare-industry-landscape-c433829b320c
How to systematically evaluate innovation:
- Commnon task framework
- Reporting guidelines
50 years of Data Science: https://www.tandfonline.com/doi/full/10.1080/10618600.2017.1384734
Common task - shared data - standard evaluation.
Tukey wrong guidelines.
Tripod Statement. https://www.tripod-statement.org/
Equator network https://www.equator-network.org/
Used for external vendors only for now. Used on data challenges.
Confessions of a pragmatic statistician. Chris Chatfield http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.93.3273&rep=rep1&type=pdf
Implentationn is the real black box.
Research on research. ImageNet for clinical data.
Innovation embedded in all departments.
Efficacy is the biggest issue. Low win value from early design. Preclinical to clinical link is the biggest win.
Clinical -> PoC as big win two. Repurposing drugs.
Qualify novel endpoints. 6 minutes walk test still used a lot.
MARINA trial with huge country variabiltiy due to number of injections required.
Ph2 LADDER trial.
Question firsts.
Combine colonoscopy and stool proteomics to avoid full anesthesia on children.
Bruno Correia https://scholar.google.ch/citations?hl=en&user=Va246xYAAAAJ&view_op=list_works&sortby=pubdate
Rethinking drug design in the artificial intelligence era https://www.nature.com/articles/s41573-019-0050-3?draft=collection
Still quite search based.
JT-VAE https://arxiv.org/pdf/1802.04364
Molecule case somehow harder as there's no alphabet and it's more 3d. (?)
Protein design: size scales as 20ˆL
Directed Evolution. Nobel prize chemistry 18. Wetlab-based, Frances Arnold. Random changes and random places in genome. Measure what one property you care about (or proxy) - fitness function. https://www.nobelprize.org/prizes/chemistry/2018/arnold/lecture/
How to do this in-silico?
-
Replace assay with predictive model
-
Replace random-greedy search with intelligent search
Sequence -> structure -> function
but can skip structure and go function! Emerging trend.
Assume there exists some predictive model - stoachastic oracle. Responses: protein expression, cell fitness. Looks like combinatorial optimisation.
Flip it. Uncertainty in predictions are now important.
ML-based protein design. DNA sequence -> protein fluoro: oracle. Add constraints, like secondary structure remains unchanged.
- Oracle (in any discrete case)
- Uncertainty
- Provide set of candidates
Brookes and Listgarten 18
Model-based optimisation: search over a distribution of proteins. Can use tools from probabilistic modelling.
Design by adaptive sampling arXiv:1810.03714. The search model could be VAE, HMM etc.
Gives probability distribution over proteins.
Kind of a first principles approach, even if NP hard.
Brookes, Park & Listgarten. ICML19 http://proceedings.mlr.press/v97/brookes19a.html
Linked to Estimation of Distribution Algorithms. https://arxiv.org/abs/1905.10474
Linked to RL.
Image generation. Nguyen et al ICCV 17.
Assumes oracle in unbiased and has good uncertainty estimate
Flip - fix desired property and see what provokes it (think Zeiler and Fergus image work). But this gives crazy looking results.
Try to fold in physics. Belonging to region ICML19.
there is no ground truth
Proposal - use GP mean model as ground truth. Then train an oracle model.
Note not Bayessian in that not acquiring new data.
11.2. U Geneva: Conditional Generation of Molecules from Disentangled Representations. Amina Mollaysa
Conditional image gen and style transfer for molecules.
SMILE -> seq model. LSTM. Brown et al 19.
molecule -> smile -> embedding -> molecule embedding concatenated with responses (?) -> LSTM. On Guacamol dataset.
No latent repn. can't do style transfer.
Developed latent model.
COnstrained ELBO
Variational Inference: A Review for Statisticians, David M. Blei, Alp Kucukelbir, Jon D. McAuliffe. https://arxiv.org/abs/1601.00670 Fixing a Broken ELBO, Alexander A. Alemi, https://arxiv.org/abs/1711.00464.
Used Grammar based encoding.
11.3. LBNL An Autoencoder for 3D Geometries of Atomic Structures with Euclidean Neural Networks, Tess E. Smidt
https://arxiv.org/abs/1802.08219
Atomic structures are hierarchy. Build in symmetry from the start.
Spherical harmonics appearing.
Can't just use scalar product due to geometry tensors.
Kapil Engel, Rossi MC, JCTC 19.
Ket of features with right symmetry properties. Cartesian coordindates of atoms don't fulfill most of these. Start with repn. Separate density for each atomic species. Need to make invariant by integrating over symmetry group. cf convolution.
Go to 3body correlations. Body order expansion equivalent to linear idea from ML approach.
Sum over atom-centred positions.
Multiscale models.
Cheng, MC PNAS 2019
DUD-E protein ligand binding. Science Advances 2017. Same molecules listed as binders and non-binders for one protein - careful!
Can predict electro densities.
Salt - sum over squares or circles gives different answers. Need global info.
Long-distance equivariant repn. Incorporating long-range physics in atomic-scale machine learning Grisafi, Andrea ; Ceriotti, Michele
Ridge regression.
11.5. Google DeepMind: AlphaFold - Improved protein structure prediction using potentials from deep learning, Andrew Senior
https://deepmind.com/blog/article/AlphaFold-Using-AI-for-scientific-discovery
Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13) https://onlinelibrary.wiley.com/doi/full/10.1002/prot.25834
Improved protein structure prediction using potentials from deep learning https://www.nature.com/articles/s41586-019-1923-7?WT.ec_id=NATURE-202001&mkt-key=005056A5C6311ED999A14F2039EAFD47&sap-outbound-id=9EB49E7379B388924AC8013686D52E221AC84B6B
Protein structure prediction.
Protein shape informs function.
Target and class of drugs.
Mitochondrial ATP Synthase MRC.
AA sequence -> 3d structure, atom coordinates.
AA residues N and 2C connected in chain to form backbone. Plus side chains, functional groups.
Structure -> function.
Experimental methods are difficult and expensive.
N residues -> 2N degrees of freedom: two angles/torsions: complete parameterisation. One representation
Can also have distance matrix repn.
Proteins can have long-range dependencies (folding).
150k entries in PDB, but very redundant. About 30k for training.
DNA constantly mutating. Inherited proteins diverge. 20N sized space. Similar sequences likely then from a common ancestor and have similar structure. Multiple sequence alignements.
Touching residues important for stability. Various instabilities exist, so a destabilising mutation may want to a stabilising co-mutation. Liely in touching residues.
FInd and align similar sequences. Compute measures of co-evolutionary correlation. Predict touching residues. Use predictions to narrow down structure search.
DeepMind - probably more info in data. Distance prediction even better than predicting contacts. Look at groups of residues -> regions. Can infer secondary structure (like strand and contact). Propagate distance constraints.
Predict histogram for distances: distogram. Predict torsions. Ramachandran plot.
Refinement and side chains from Rosetta.
Inductive bias - using a 2d CNN.
Deep dilated convolutional residual network.
Cropping - O(Lˆ2) memory needed for all distance pairs. Crop to 64x64 squares of distance matrix. Predict also interblocks to get long range interactions. Fits in memory. Works like data augmentation, using eg strides. Centre-weighted the predictions to avoid edge effects.
Predictions are on blocks that are then combine. Can Ensemble too. Edges have.
- random crop
- Add noise
- resample multiple sequences.
- lr decay
- multiple networks with different ICs ensembled.
Nˆ2 distances and only 2N dof!
Potentials for various terms - distance + torsion + no-touching.
Can initialize with a particular secondary structure.
There are different domains - different folding for different parts. Can cut up, but not used in sgd approach with sklearn sgd.
Approaching biological relevance.
Interpretability limited, but being worked on.
Code open-sourced.
ProSPr: Democratized Implementation of Alphafold Protein Distance Prediction Network https://www.biorxiv.org/content/10.1101/830273v1
https://github.com/dellacortelab/prospr
Expectation that Transformers will be appearing in next CASP.
11.7. LabGenius - Knitting together synthetic biology, machine learning and robotics, Katya Putintseva
20k human body proteins.
20ˆ27000 - largest protein.
Not all seen in nature. Some have also 'disappeared'
Each point a protein variant. Proximity means sequence is close.
Build a map (?) -> empirical computation: experiments and computers.
- build trillions of protein variants
- test for what you want
- learn - ML mode
- Design new
- Continue
Multi response. Deconvolution. Test for each property. Multi-objective optimisation.
Developing a protein embedding space.
Using quality score from NGS.
https://github.com/epfl-exts/amld20-anomaly-detection
https://github.com/epfl-exts/amld20-text-classification
https://github.com/amueller/introduction_to_ml_with_python
Standard regex and logistic regression in sklearn pipeline https://scikit-learn.org/stable/modules/generated/sklearn.pipeline.Pipeline.html
https://github.com/epfl-exts/amld20-image-classification
Some nice basics with histograms etc., then CNN.
https://github.com/epfl-exts/amld20-image-classification
https://www.whattolabel.com (AMLD2020)
GIOTTO: https://github.com/giotto-ai/giotto-tda
NLP tutorial from DSSS19: https://github.com/steve-wilson/ds32019
Causality link from Kostas (met at DSSS18, now at Roche): https://www.edx.org/bio/miguel-hernan