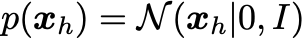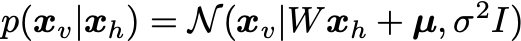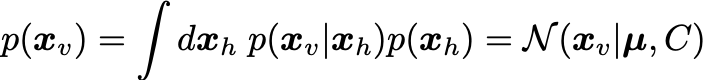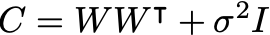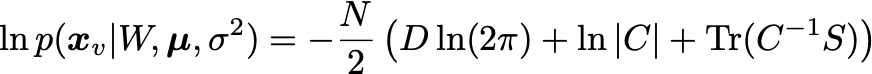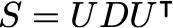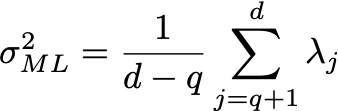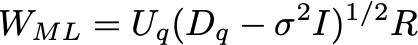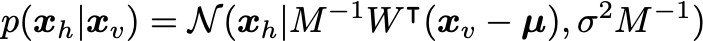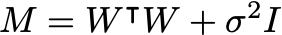You can read a complete tutorial on Medium here.
- Python:
python prob_pca.py. The figures are output to the figures_py directory. - Mathematica: Run the notebook
prob_pca.nb. The figures are output to the figures_ma directory.
You can find more information in the original paper: "Probabilistic principal component analysis" by Tipping & Bishop.
Let's import and plot some 2D data:
data = import_data()
d = data.shape[1]
print("\n---\n")
mu_ml = np.mean(data,axis=0)
print("Data mean:")
print(mu_ml)
data_cov = np.cov(data,rowvar=False)
print("Data cov:")
print(data_cov)
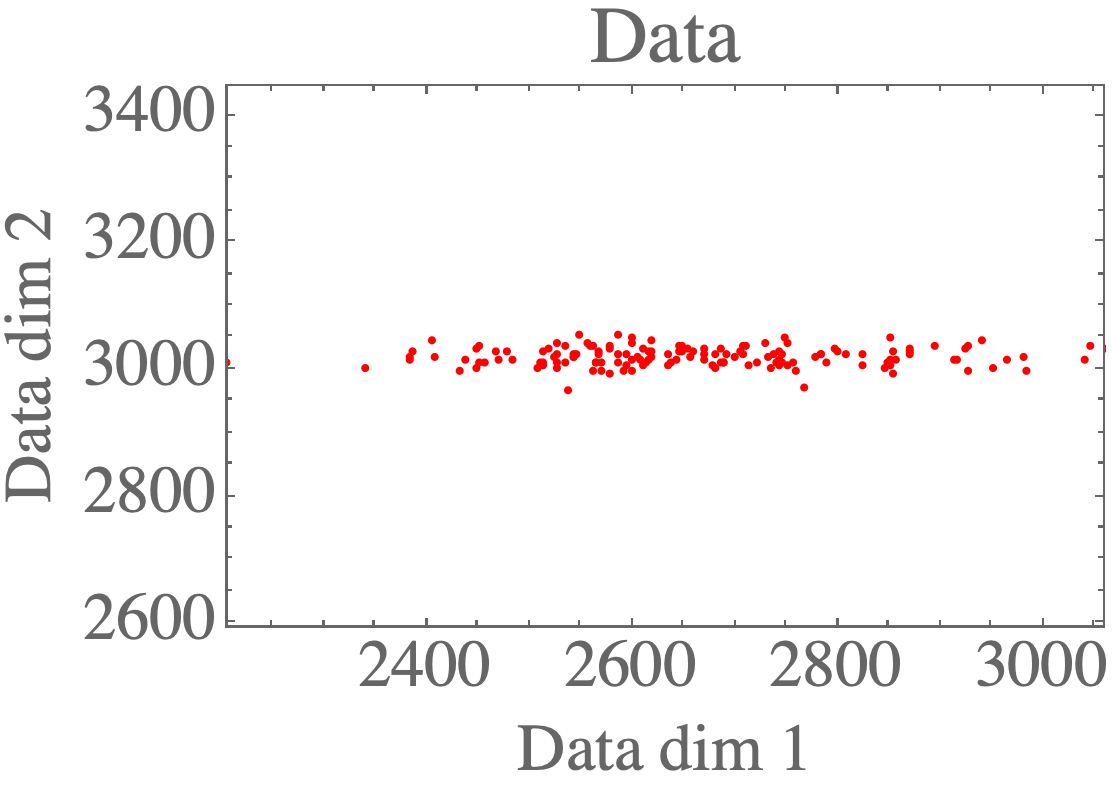
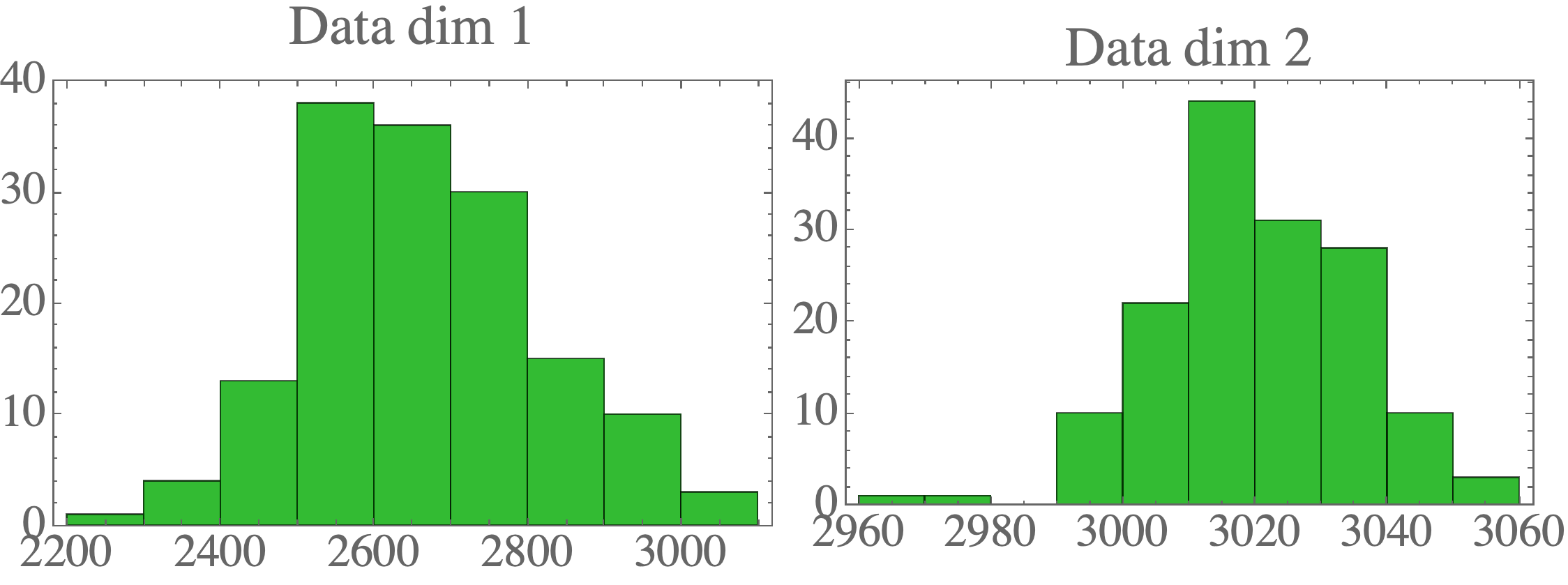
You can see the plotting functions in the complete repo. Visualizing the data shows the 2D distribution:
Assume the latent distribution has the form:
The visibles are sampled from the conditional distribution:
From these, the marginal distribution for the visibles is:
We are interested in maximizing the log likelihood:
where S is the covariance matrix of the data. The solution is obtained using the eigendecomposition of the data covariance matrix S:
Where the columns of U are the eigenvectors, and D is a diagonal matrix with the eigenvalues \lambda. Let the number of dimensions of the dataset be d (in this case, d=2). Let the number of latent variables be q, then the maximum likelihood solution is:
and \mu are just set to the mean of the data.
Here the eigenvalues \lambda are sorted from high to low, with \lambda_1 being the largest and \lambda_d the smallest. Here U_q is the matrix where columns are the corresponding eigenvectors of the q largest eigenvalues, and D_q is a diagonal matrix with the largest eigenvalues. R is an arbitrary rotation matrix - here we can simply take R=I for simplicity (see Bishop for a detailed discussion). We have discarded the dimensions beyond q - the ML variance \sigma^2 is then the average variance of these discarded dimensions.
Here is the corresponding Python code to calculate these max-likelihood solutions:
# No hidden variables < no visibles = d
q = 1
# Variance
lambdas, eigenvecs = np.linalg.eig(data_cov)
idx = lambdas.argsort()[::-1]
lambdas = lambdas[idx]
eigenvecs = - eigenvecs[:,idx]
print(eigenvecs)
# print(eigenvecs @ np.diag(lambdas) @ np.transpose(eigenvecs))
var_ml = (1.0 / (d-q)) * sum([lambdas[j] for j in range(q,d)])
print("Var ML:")
print(var_ml)
# Weight matrix
uq = eigenvecs[:,:q]
print("uq:")
print(uq)
lambdaq = np.diag(lambdas[:q])
print("lambdaq")
print(lambdaq)
weight_ml = uq * np.sqrt(lambdaq - var_ml * np.eye(q))
print("Weight matrix ML:")
print(weight_ml)
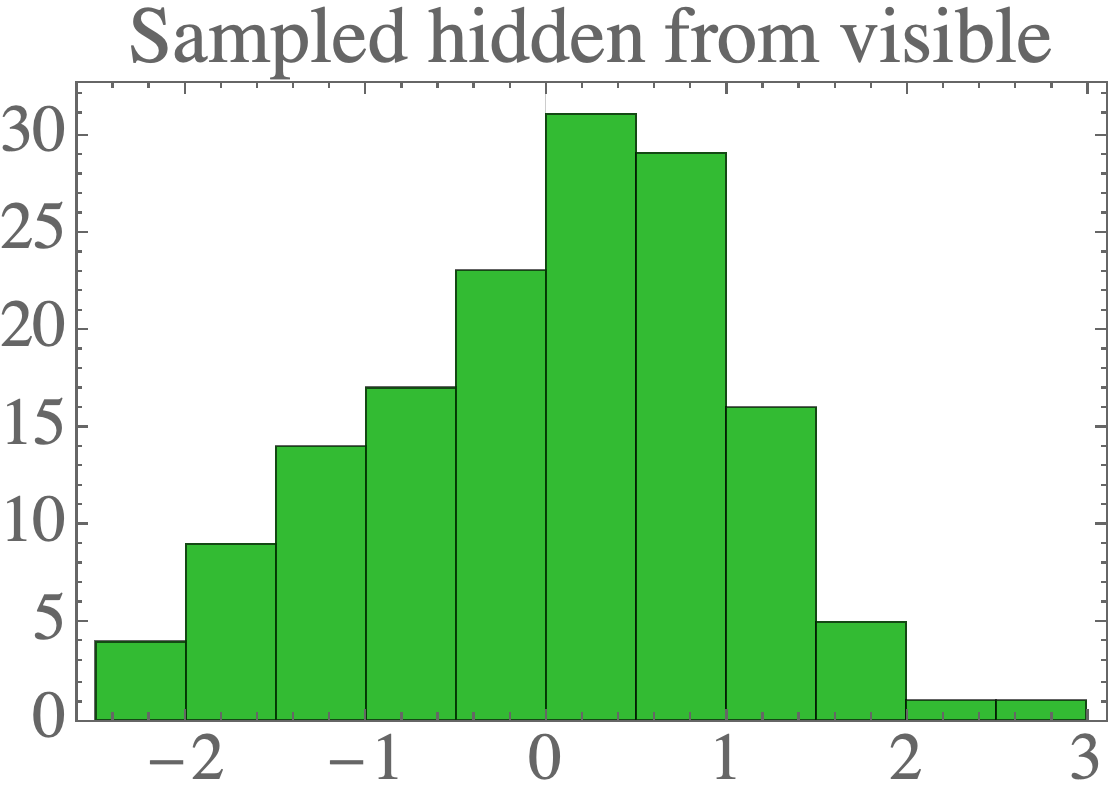
After determining the ML parameters, we can sample the hidden units from the visible according to:
You can implement it in Python as follows:
act_hidden = sample_hidden_given_visible(
weight_ml=weight_ml,
mu_ml=mu_ml,
var_ml=var_ml,
visible_samples=data
)
where we have defined:
def sample_hidden_given_visible(
weight_ml : np.array,
mu_ml : np.array,
var_ml : float,
visible_samples : np.array
) -> np.array:
q = weight_ml.shape[1]
m = np.transpose(weight_ml) @ weight_ml + var_ml * np.eye(q)
cov = var_ml * np.linalg.inv(m)
act_hidden = []
for data_visible in visible_samples:
mean = np.linalg.inv(m) @ np.transpose(weight_ml) @ (data_visible - mu_ml)
sample = np.random.multivariate_normal(mean,cov,size=1)
act_hidden.append(sample[0])
return np.array(act_hidden)
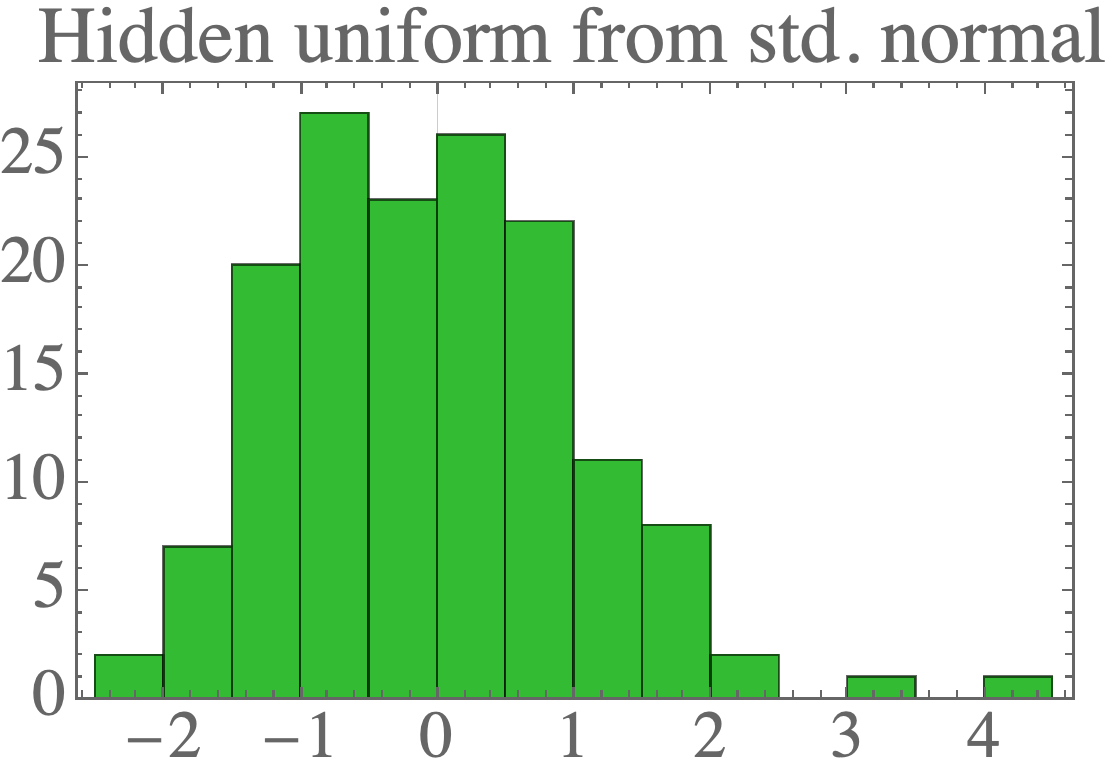
The result is data which looks a lot like the standard normal distribution:
We can sample new data points by first drawing new samples from the hidden distribution (a standard normal):
mean_hidden = np.full(q,0)
cov_hidden = np.eye(q)
no_samples = len(data)
samples_hidden = np.random.multivariate_normal(mean_hidden,cov_hidden,size=no_samples)
and then sample new visible samples from those:
act_visible = sample_visible_given_hidden(
weight_ml=weight_ml,
mu_ml=mu_ml,
var_ml=var_ml,
hidden_samples=samples_hidden
)
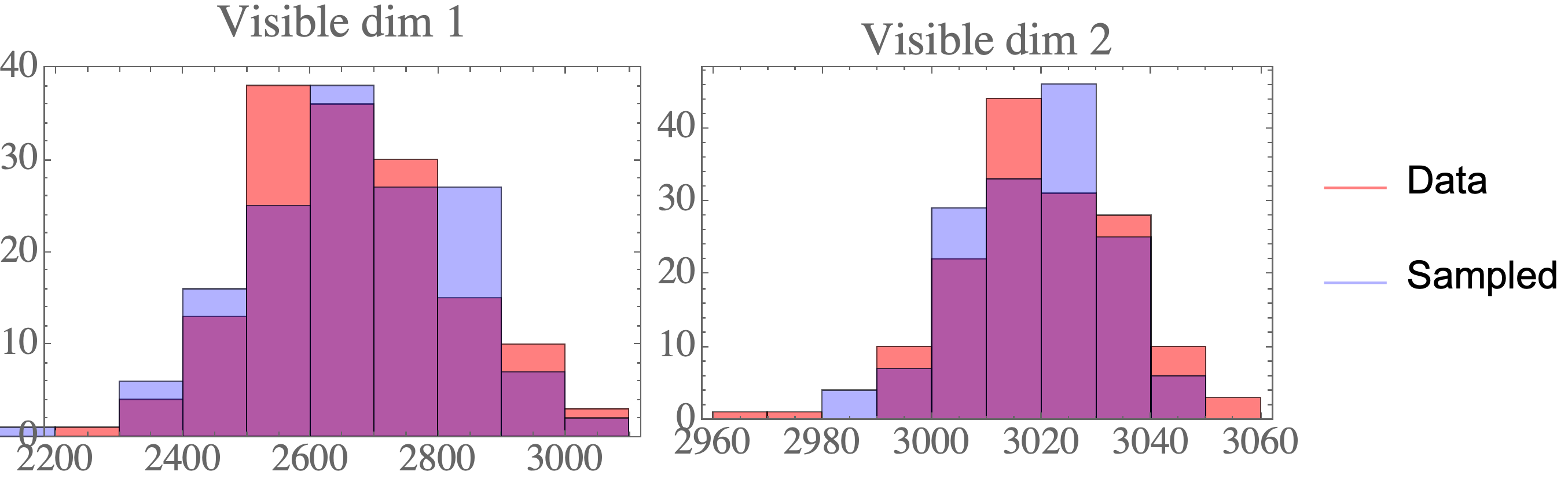
print("Covariance visibles (data):")
print(data_cov)
print("Covariance visibles (sampled):")
print(np.cov(act_visible,rowvar=False))
print("Mean visibles (data):")
print(np.mean(data,axis=0))
print("Mean visibles (sampled):")
print(np.mean(act_visible,axis=0))
where we have defined:
def sample_visible_given_hidden(
weight_ml : np.array,
mu_ml : np.array,
var_ml : float,
hidden_samples : np.array
) -> np.array:
d = weight_ml.shape[0]
act_visible = []
for data_hidden in hidden_samples:
mean = weight_ml @ data_hidden + mu_ml
cov = var_ml * np.eye(d)
sample = np.random.multivariate_normal(mean,cov,size=1)
act_visible.append(sample[0])
return np.array(act_visible)
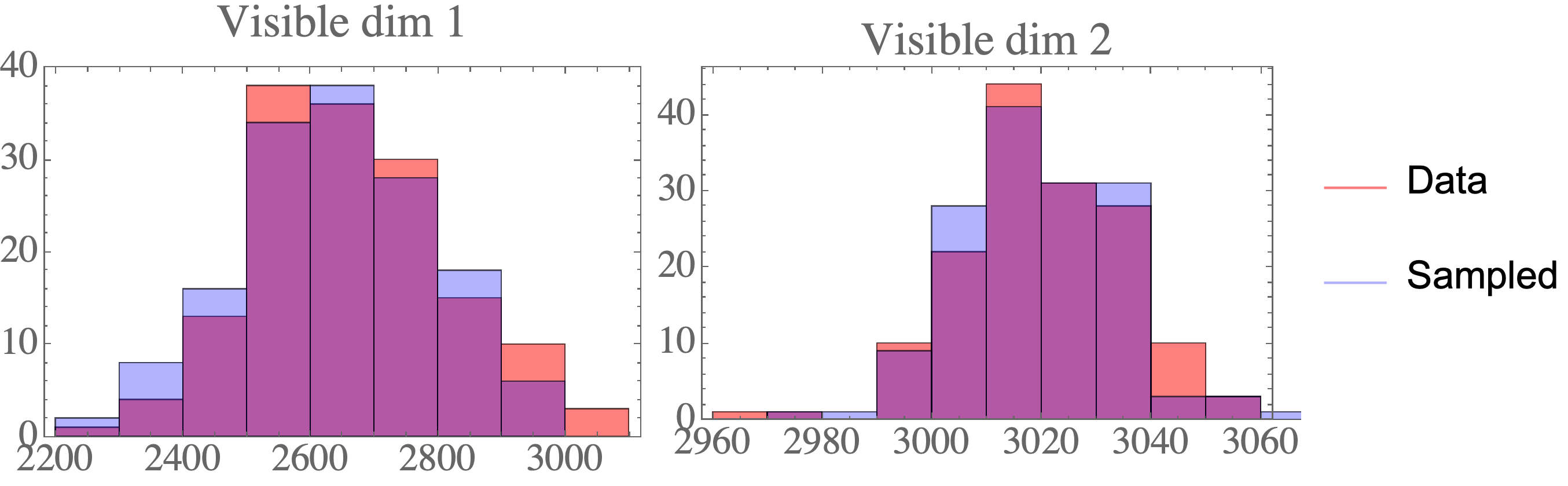
The result are data points that closely resemble the data distribution:
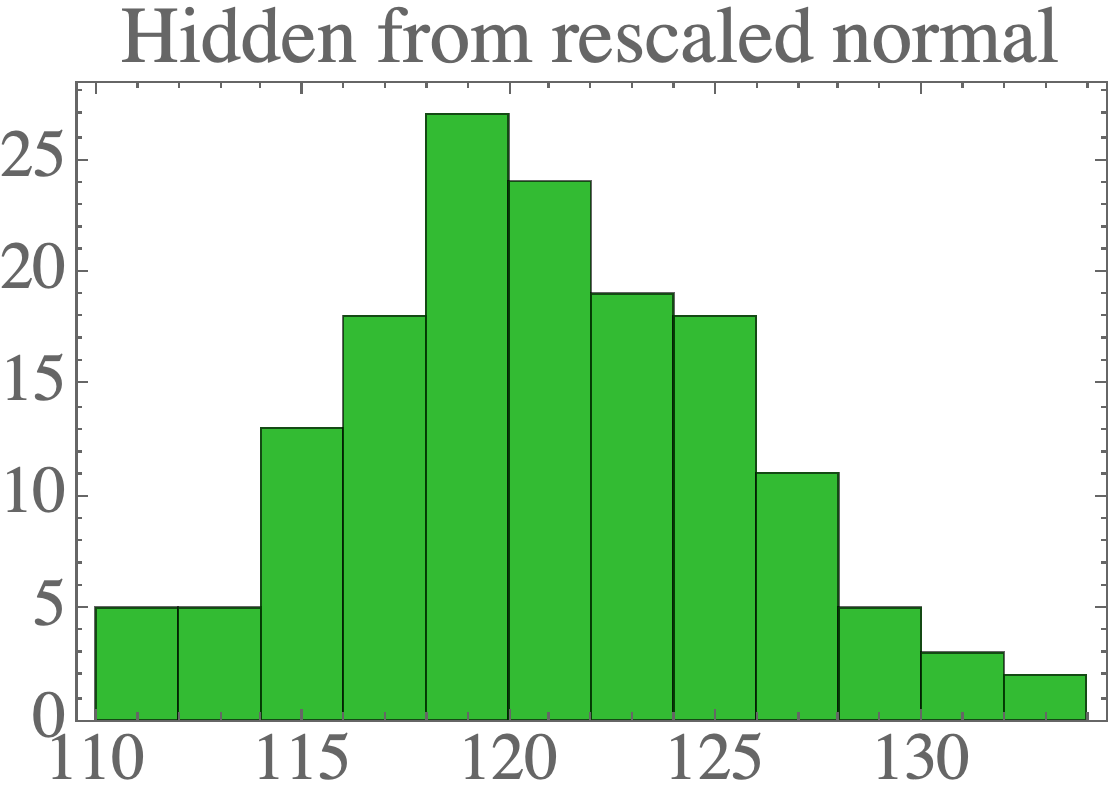
Finally, we can rescale the latent variables to have any Gaussian distribution:
For example:
mean_hidden = np.array([120.0])
cov_hidden = np.array([[23.0]])
no_samples = len(data)
samples_hidden = np.random.multivariate_normal(mean_hidden,cov_hidden,size=no_samples)
We can simply transform the parameters and then still sample new valid visible samples from those:
Nota that \sigma^2 is unchanged from before.
In Python we can do the rescaling:
weight_ml_rescaled = weight_ml @ np.linalg.inv(sl.sqrtm(cov_hidden))
mu_ml_rescaled = mu_ml - weight_ml_rescaled @ mean_hidden
print("Mean ML rescaled:")
print(mu_ml_rescaled)
print("Weight matrix ML rescaled:")
print(weight_ml_rescaled)
and then repeat the sampling with the new weights & mean:
act_visible = sample_visible_given_hidden(
weight_ml=weight_ml_rescaled,
mu_ml=mu_ml_rescaled,
var_ml=var_ml,
hidden_samples=samples_hidden
)
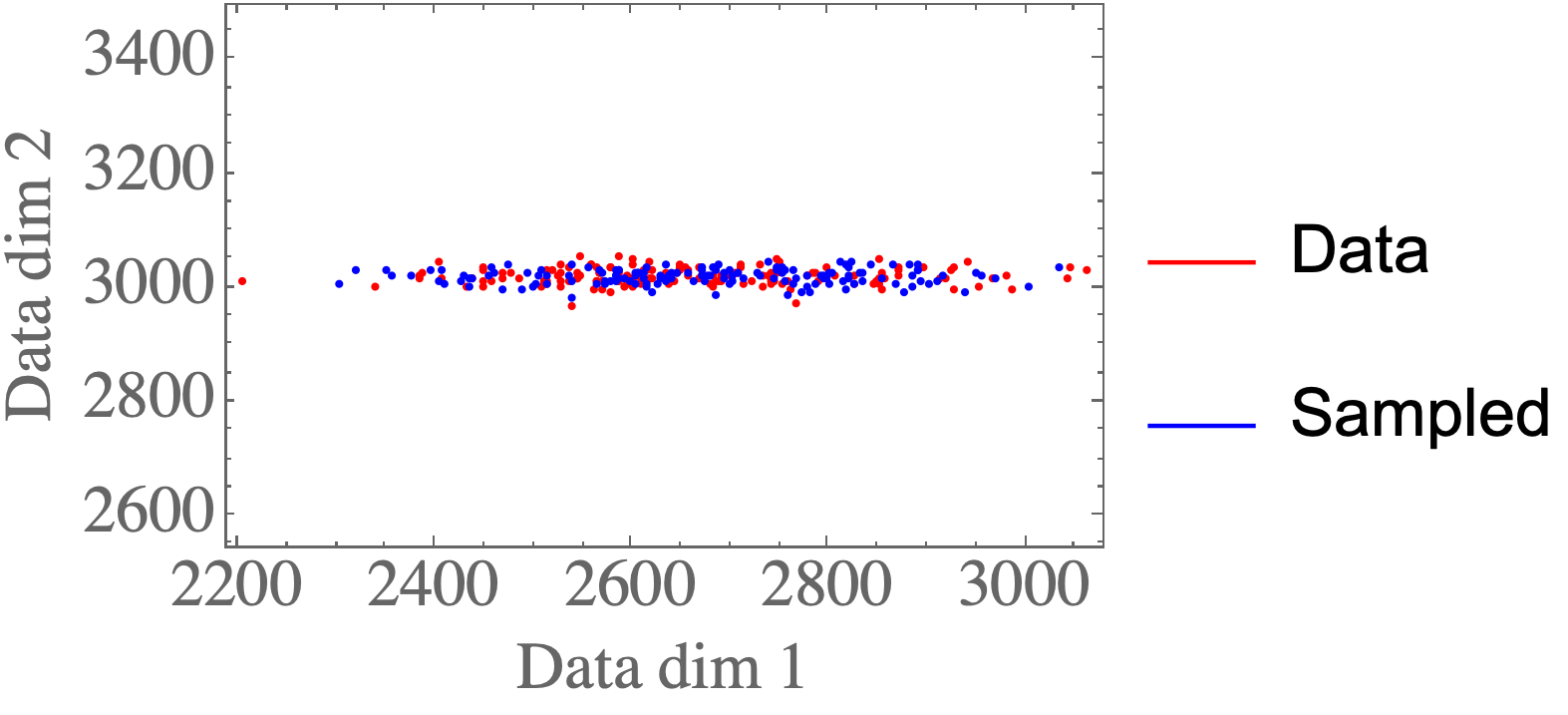
Again, the samples look like they could come from the original data distribution: