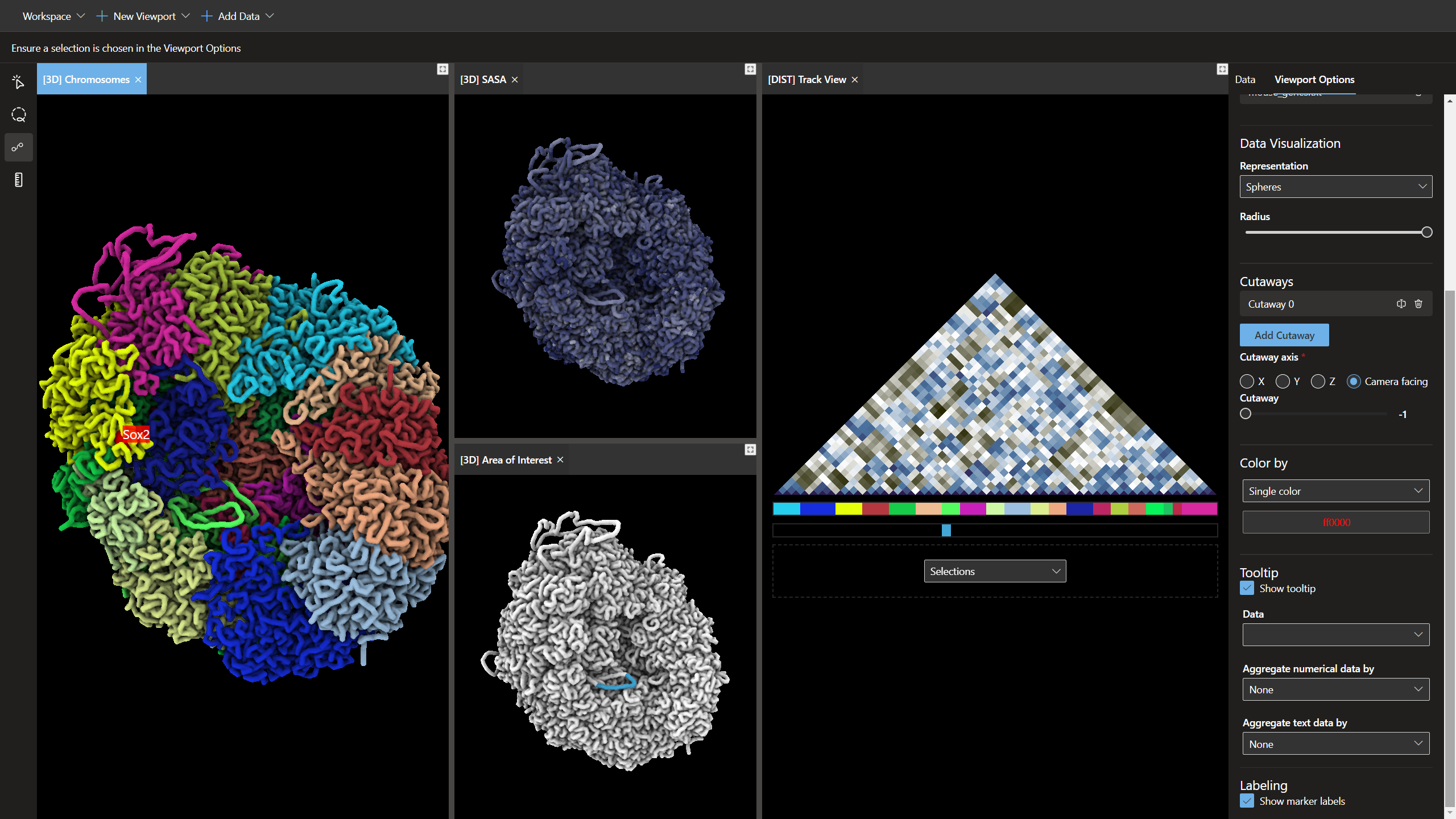
Chromoskein is a tool for visualizing genomic data with a special focus 3D chromatin structures predicted from Hi-C.
The project uses WebGPU which is currently under development. In order to run Chromoskein, you will need experimental versions of modern browsers. We recommend Google Chrome Canary.
Chromoskein is developed at VisitLab from Masaryk University, Brno, Czech Republic.
Windows 10 (Creator's update and later):
-
Enable 'Developer mode'
-
Clone with symlinks enabled
git clone -c core.symlinks=true
Run dev server
cd appnpm run serve
- Google Chrome Canary
- Enable Unsafe WebGPU flag:
chrome://flags/#enable-unsafe-webgpu - Make sure hardware acceleration is enabled:
chrome://settings/system> Use hardware acceleration when available- Graphics status can be checked in:
chrome://gpu
- Graphics status can be checked in:
- Chromoskein on macOS refreshes when data is loaded in 3D viewport? Disable Back-forward cache flag:
chrome://flags/#back-forward-cache. This is probably a bug in Chrome and the flag won't be needed in the future.
- Enable Unsafe WebGPU flag:
Exemplary deployed version of ChromoSkein is available at https://visitlab.pages.fi.muni.cz/chromatin/chromoskein/
A demo workspace with sample dataset can be found under Workspace > Open exemplary workspace