This page describes the method to construct robust persistence diagrams, as implemented in our paper Robust Persistence Diagrams using Reproducing Kernels.
You will need the following dependencies for implementing the analyses using R. Please run the following code:
pkgs <- c("dplyr","plotrix","spatstat","TDA","hitandrun","functional","Rfast","plotly","viridis","plot3D")
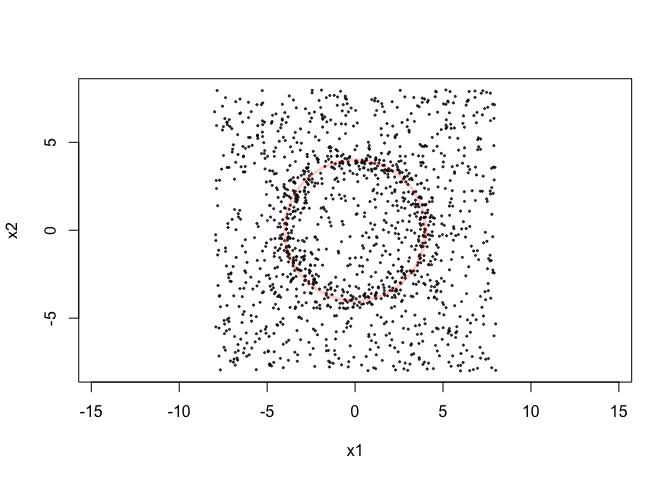
sapply(pkgs, install.packages, character.only=TRUE)We start by sampling points 𝕏n from a circle in 2D with some uniform noise in the enclosing region.
set.seed(2020)
signal <- circleUnif(400,4)*rnorm(400,1,0.1)
noise <- matrix(runif(2*1000,-8,8),ncol=2)
X <- rbind(signal,noise)
plot(X,asp=1,pch=20,cex=0.4,col=alpha("black",0.7))
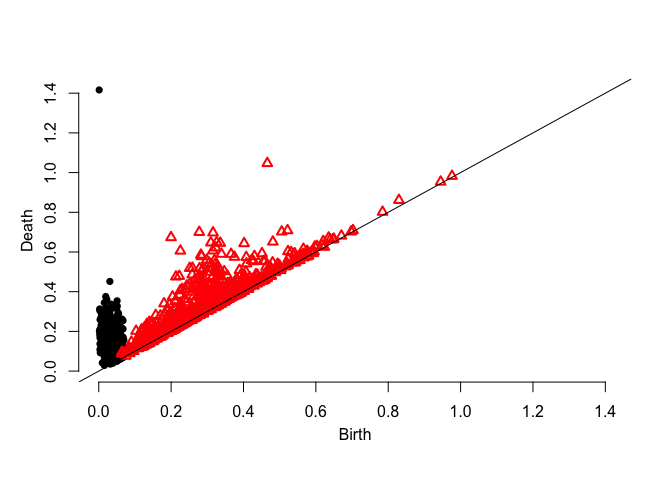
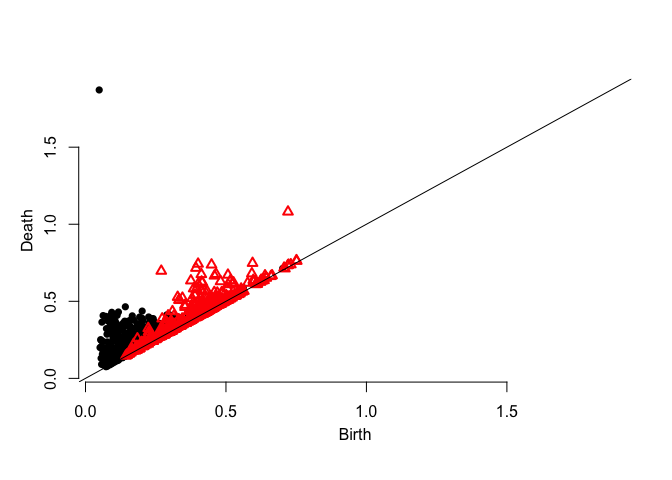
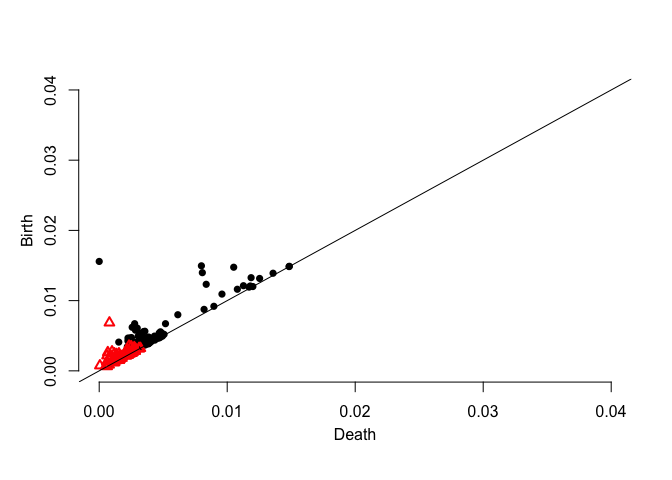
draw.circle(0,0,4,border = alpha('red',0.2),lwd=3)Here are the persistence diagrams.
dgm.distFct <- ph.distfun(X,by=0.1,H=0)$diagram
plot.diagram(dgm.distFct)dgm.dtm <- ph.dtm(X,by=0.1,m0 = M0(X,3))$diagram
plot.diagram(dgm.dtm)dgm.kde <- ph.kde(X,by=0.1,H=bw(X,3))$diagram
plot.diagram(dgm.kde)dgm.rkde <- ph.rkde2(X,by=0.1,H=bw(X,3))$diagram
plot.diagram(dgm.rkde)