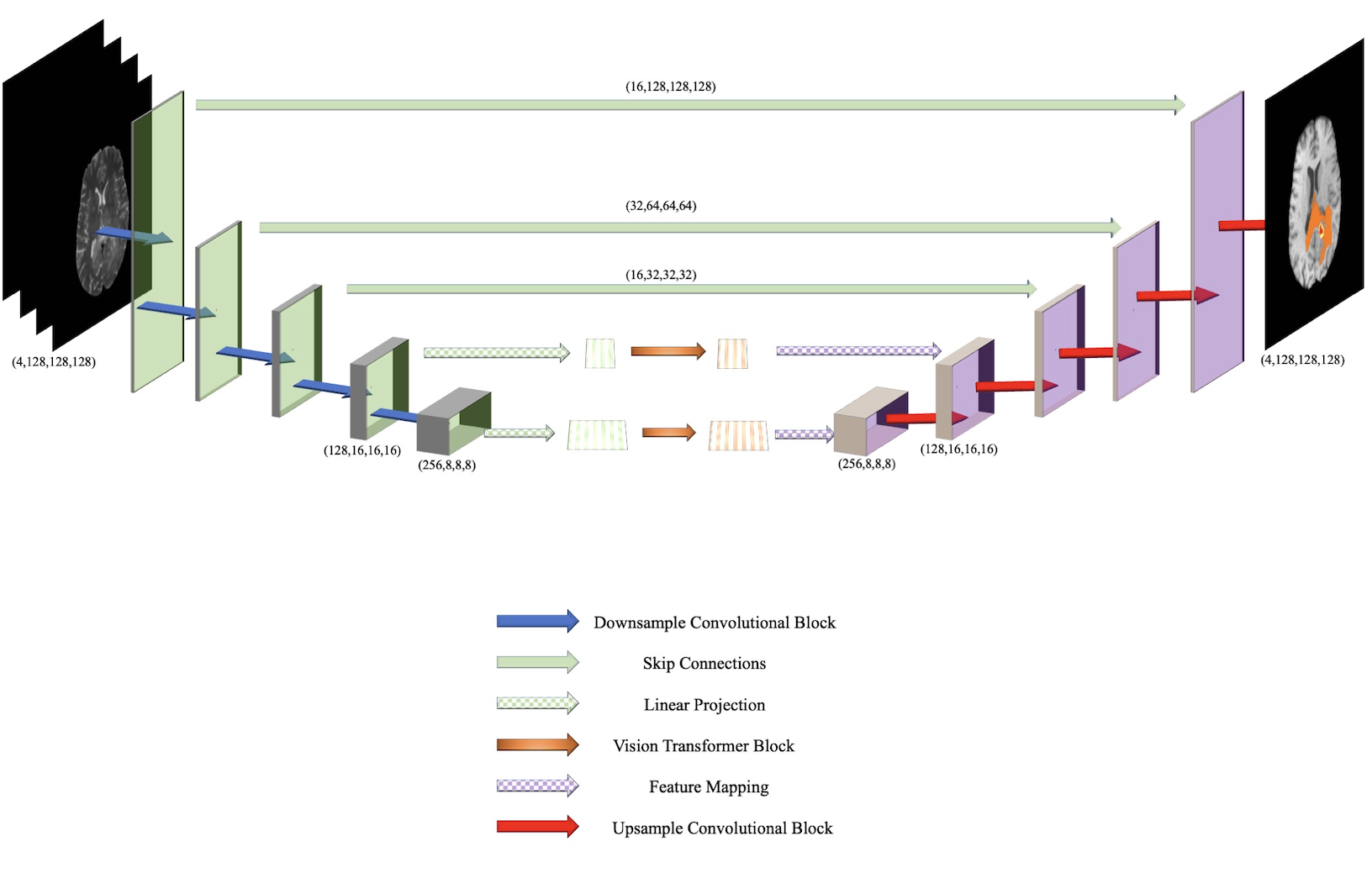
This repo is the source code for BiTr-Unet: a CNN-Transformer Combined Network for MRI Brain Tumor Segmentation. The dataset that this model is designed for is BraTS 2021, which can be retrieved from https://www.synapse.org/#!Synapse:syn25829067/wiki/611501
- python 3.7
- pytorch 1.6.0
- torchvision 0.7.0
- pickle
- nibabel
- SimpleITK
- imageio
Please prepare an environment with python=3.7, and then use the command "pip install -r requirements.txt" for the dependencies.
Mount the folders of BraTS 2021 training and validation dataset respectively under the folder "data". Modify path and Run "generate_train_list.py" and "generate_validation_list.py" to generate the train.txt and valid.txt, which are required for the next steps. Unzip "output_size_template.zip" and keep the unzipped file where it is, which is worked as a reference for the model to automatically output the segmentation with a proper size.
python3 generate_train_list.py
python3 generate_valid_list.py
unzip output_size_template.zip
Here is an example illustrating the proper way to mount the BraTS 2021 dataset: "./data/BraTS2021_TrainingData/case_ID/case_ID_flair.nii.gz" The generated train.txt should be moved to "./data/BraTS2021_TrainingData/".
Modify path and Run "preprocess.py" to generate a pkl file for every case within its case_ID folder, which are required for the next steps.
python3 preprocess.py
Modify path and Run "train.py" :
python3 -m torch.distributed.launch --nproc_per_node=4 --master_port 20003 train.py
Modify path and Run "test.py" :
python3 test.py
Mount your output folder of nii.gz files under the postprocess folder. Modify path and Run "ensemble_by_majority_voting.py" or "connected_components.py" for two different postprocessing methods.
python3 ensemble_by_majority_voting.py
python3 connected_components.py
Use the evaluation folder to calculation the Dice Score of the segmentation of the validation data.
1.TransBTS 2.two-stage-VAE-Attention-gate-BraTS2020 3.open_brats2020 4.nnUNet
Q. Jia, H. Shu (2022). "BiTr-Unet: a CNN-Transformer Combined Network for MRI Brain Tumor Segmentation". Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, LNCS 12963, pp. 3–14. https://doi.org/10.1007/978-3-031-09002-8_1