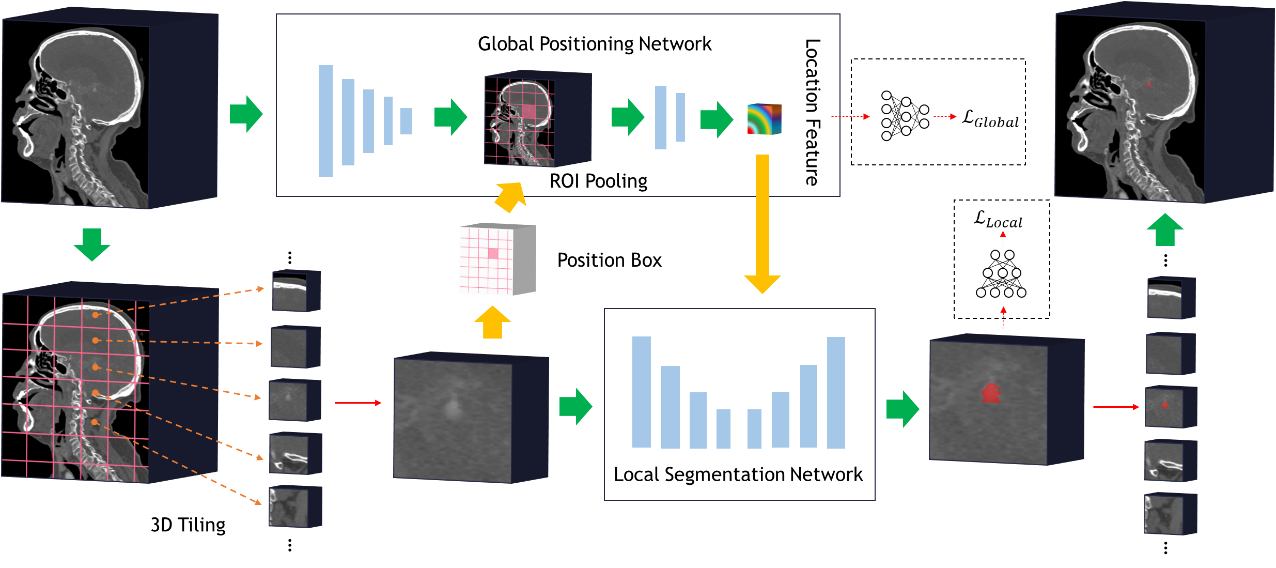
GLIA-Net: Global Localization based Intracranial Aneurysm Network
This is the official repository of GLIA-Net, a segmentation network for intracranial aneurysm on CTA images using pytorch. If you find it useful in your research, please cite our paper: https://doi.org/10.1016/j.patter.2020.100197
License
The code is licensed for non-commerical academic research purpose only.
Requirements
The project has been tested in both Windows 10 and Ubuntu 16.04 using python 3.6. But the training and evaluation code cannot use multi-processing on windows. Other packages can be installed using pip or conda.
- torch>=1.4
- torchvision
- tensorboardX
- medpy
- SimpleITK>=2.0.0rc1
- PyYAML
- pillow
- colorlog
Prepare data
If you want to train your model, put the data in medical_data/ and
prepare an instance information file like medical_data/aneurysm_seg.csv.
All the images should be in nii.gz format.
Train
Prepare a training configuration file like configs/GLIA-Net.yaml.
Then run
python train.py -c GLIA-Net
The logs, checkpoints, summaries and other running assets will be saved to exp/.
Or you can download a pretrained model - checkpoint-0245700.pt.
Then put it to exp/GLIA-Net/1/ckpt.
Evaluation
Prepare an evaluation configuration file like configs/eval_GLIA-Net.yaml,
which should have a filename prefix of 'eval_'.
Then run
python evaluate.py -c eval_GLIA-Net
Not that this is an evaluation on patches, which depends on how to generate all patches. If you want to evaluate on whole cases, run the inference code to generate predictions and then
python evaluate_per_case.py \
-c eval_per_case \
-g path/to/ground_truth \
-p path/to/predictions
View the tensorboard
You can view the training and evaluation information using tensorboard.
Run tensorboard --logdir=exp to run a tensorboard server.
Inference
Prepare an inference configureation file like configs/inference_GLIA-Net.yaml,
which should have a filename prefix of 'inference_'.
Then run
python inference.py \
-c inference_GLIA-Net \
-i path/to/input/file/or/folder \
-t nii
-o path/to/save/predictions
The inference code support single or multiple input images and image format of nii.gz or dicom series. If you want to refine the predictions, run the code
python refine_segmenation.py \
-i path/to/predictions/before/refine \
-o path/to/save/refined/predictions
Advanced functions
The code support other functions like different devices by setting -d to "cpu", "0", or "1,2,3"...,
multi-thread and multi-process data pipeline, and so on.
You can refer to the input argument instructions in train.py, evaluation.py and inference.py.
You can also modify the configurations in configs/.
The training code using batch_size of 1 shall require about 3GB of memory. You can increase the batch size according to your memory capacity.