Enhancing MR imaging driven Alzheimer’s disease classification performance using generative adversarial learning
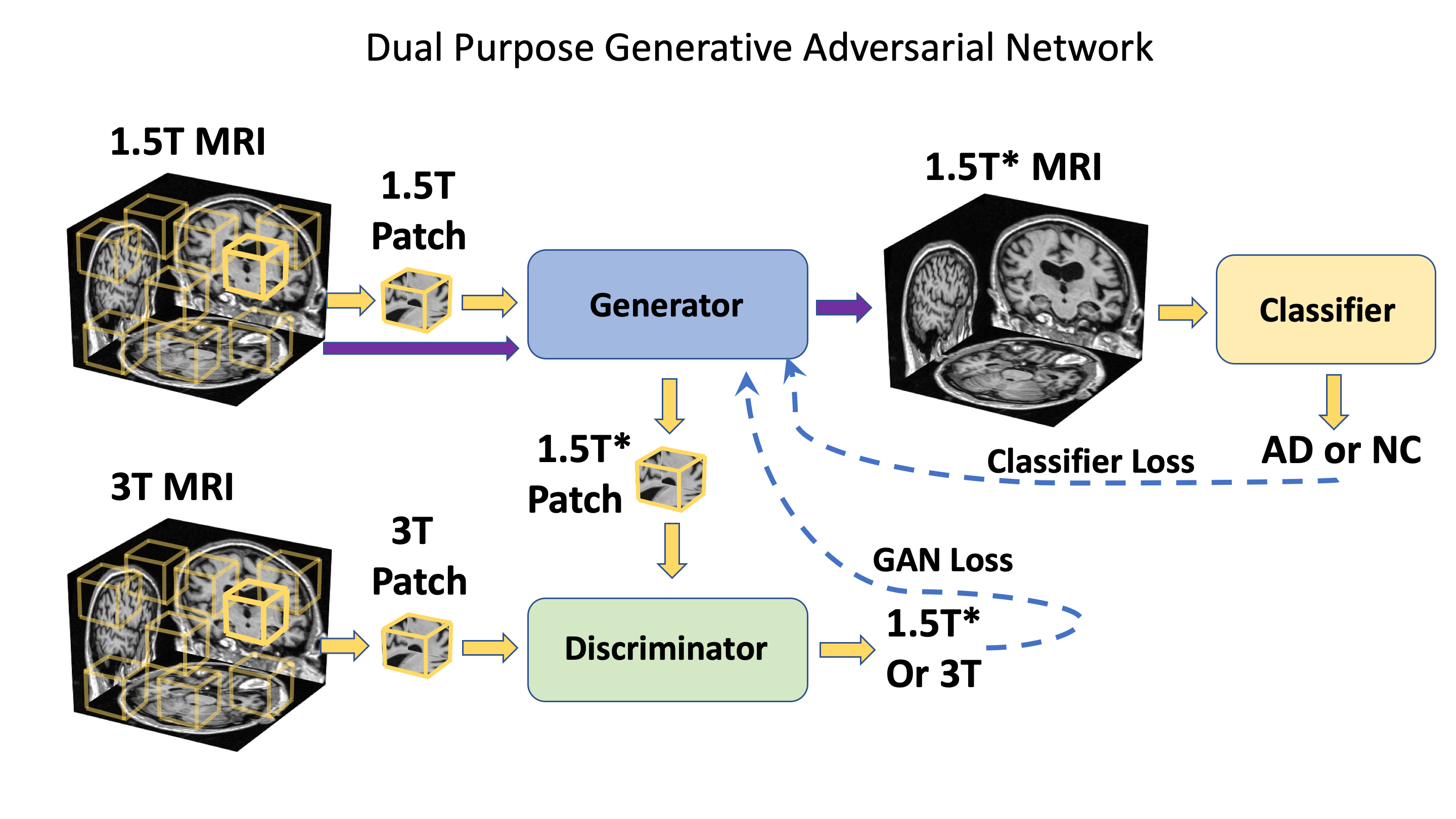
This repo contains a PyTorch implementation of a deep learning framework that enhance Alzheimer’s disease (AD) classification performance using MRI scans of multiple magnetic field strengths, while also improve the image quality. The framework contains a generative adversarial network (GAN), a fully convolutional networks (FCN), and a multilayer perceptron. See below for the overall structure of our framework.
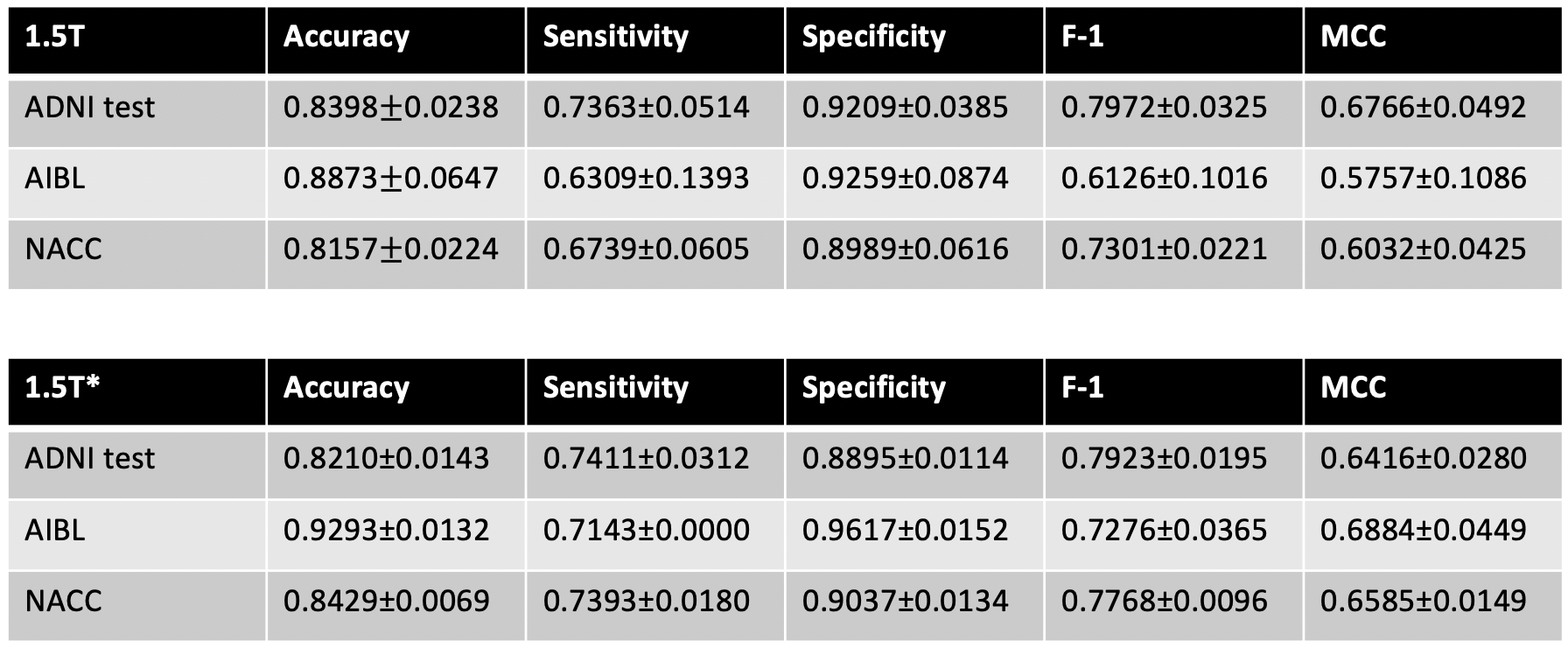
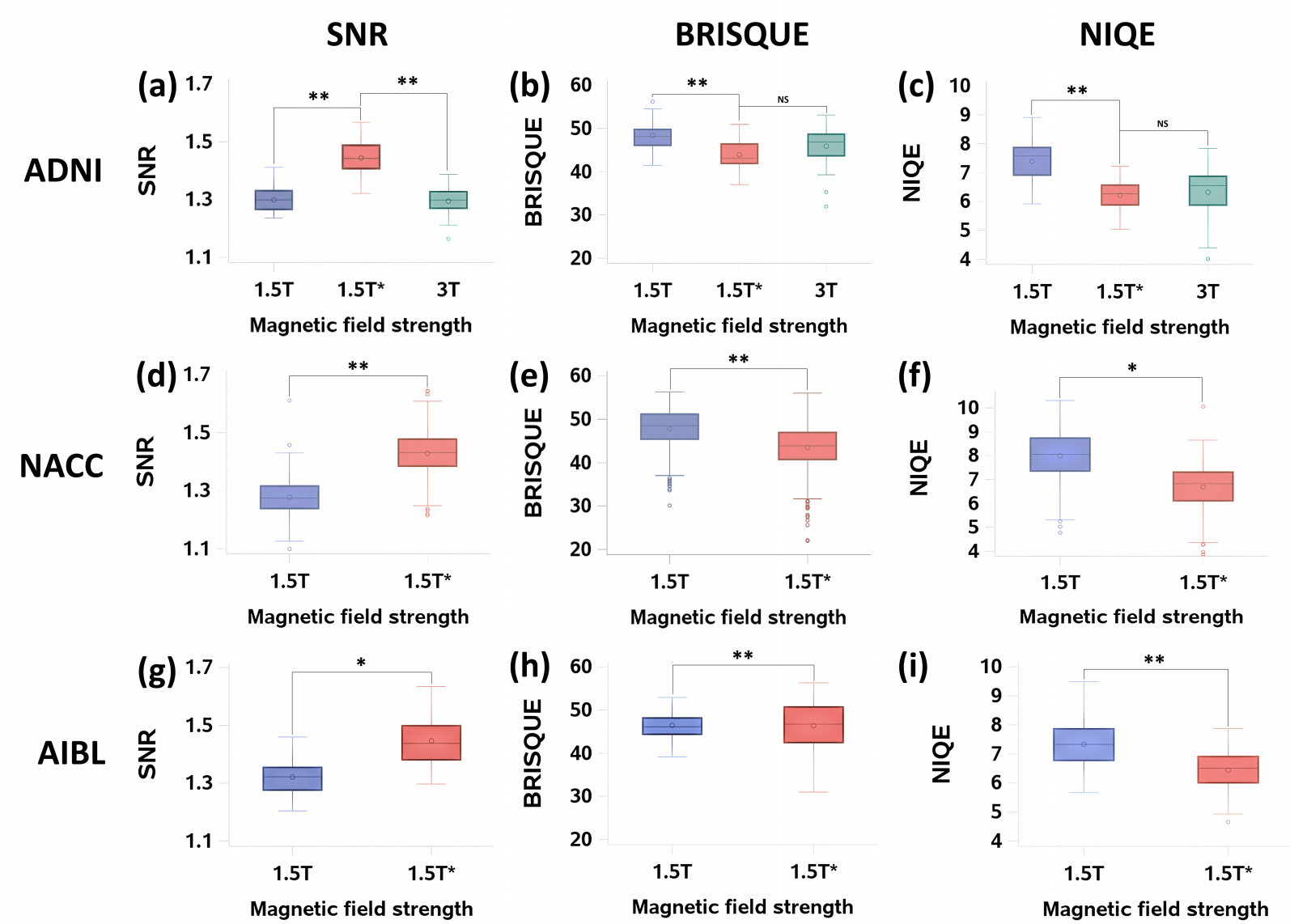
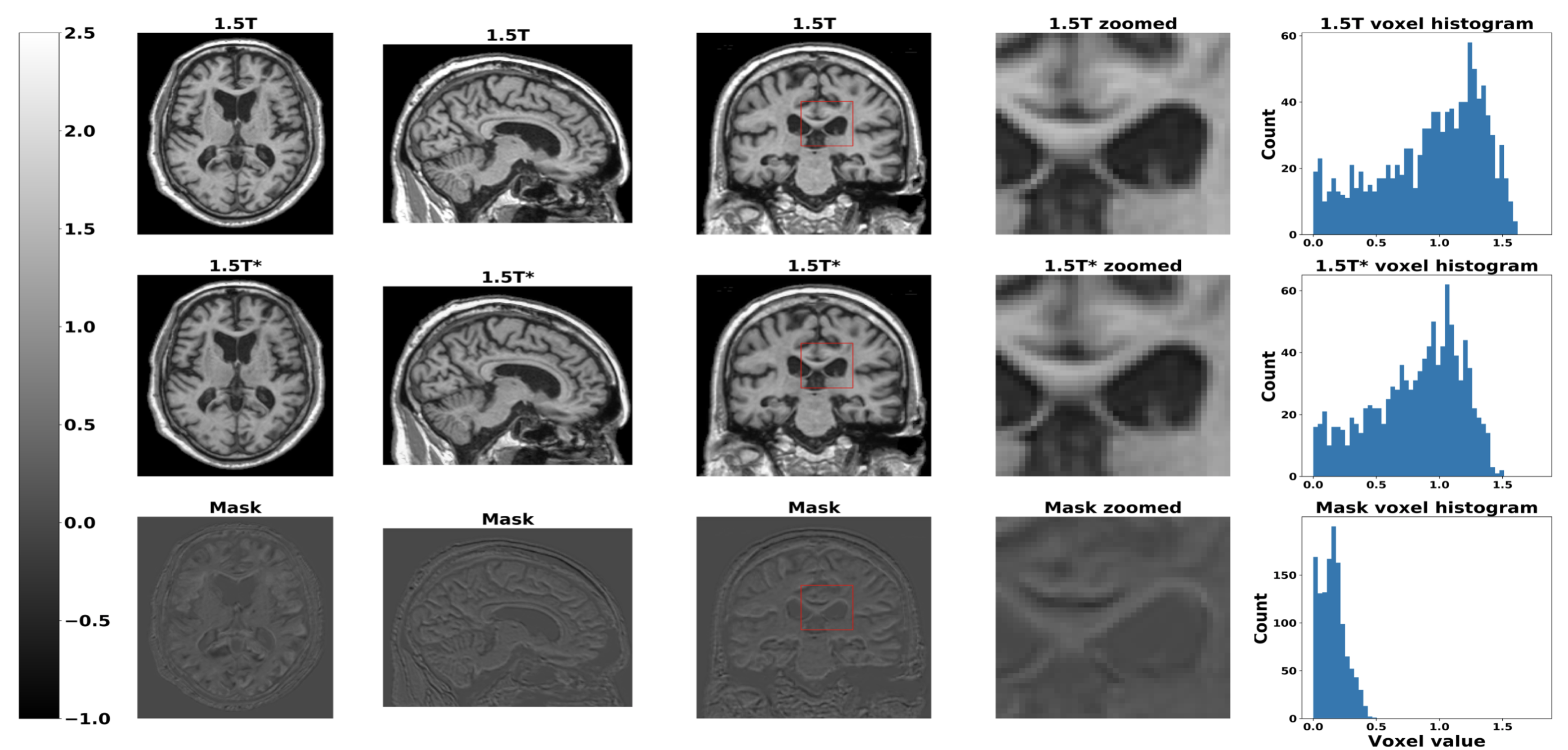
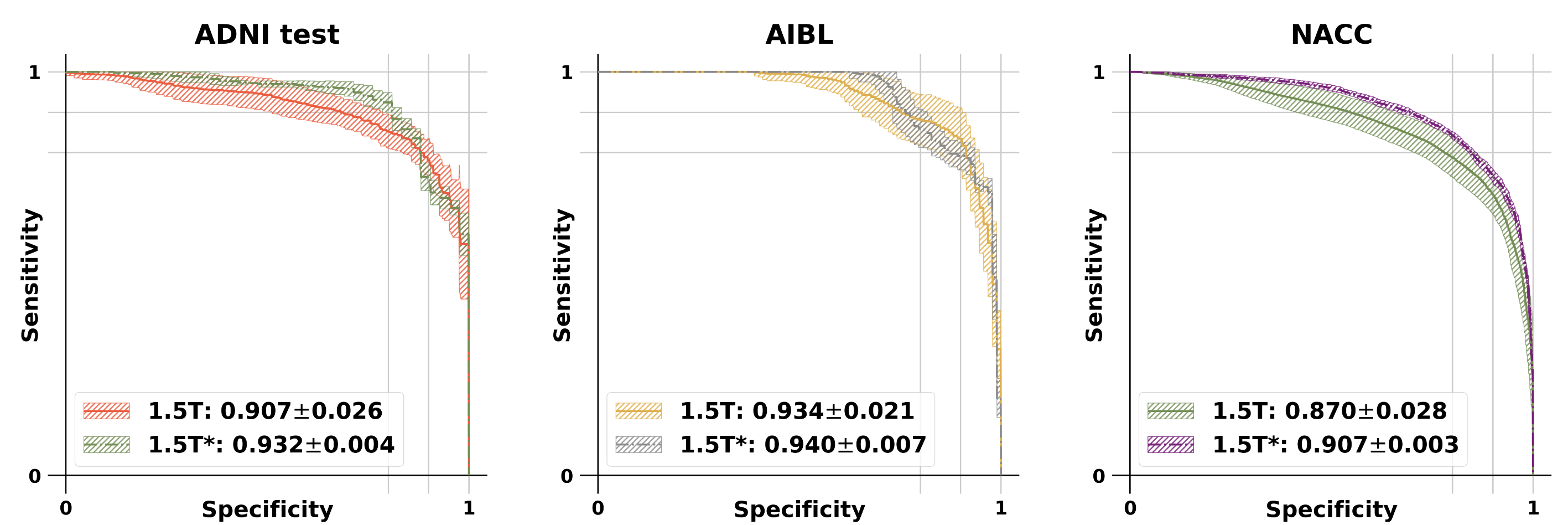
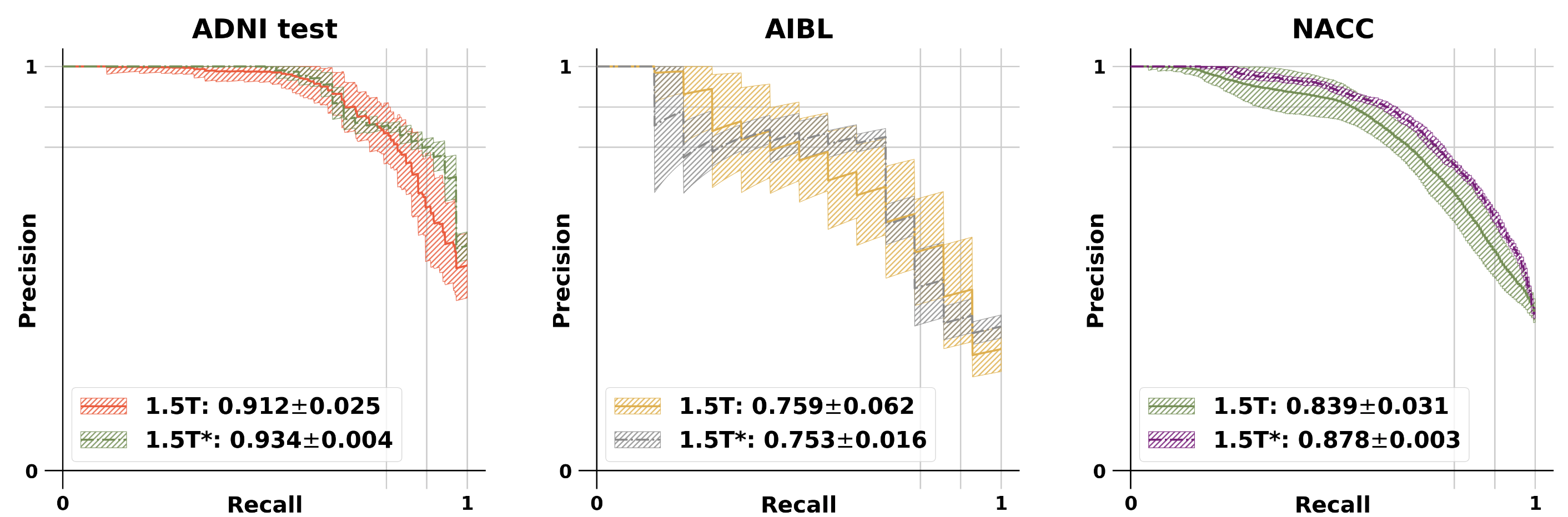
The modified GAN was developed on ADNI training and validation sets and its performance was evaluated on ADNI testing set and 2 external testing datasets (NACC and AIBL). See below for the results.
Model performance:
Image quality metrics:
Sample output:
ROC curves:
Please refer to our paper for more details.
These instructions will help you properly configure and use the tool.
We trained, validated and tested the framework using the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset. To investigate the generalizability of the framework, we externally tested the framework on the National Alzheimer's Coordinating Center (NACC) and the Australian Imaging Biomarkers and Lifestyle Study of Ageing (AIBL) datasets.
To download the raw data, please contact those affiliations directly. In "./lookupcsv/" folder, we provided csv table containing subjects details used in this study for each dataset. We also provided all data preprocessing manuscripts in "./Data_Preprocess/" folder. After data preprocssing, the data can be stored in the folder structure like below:
data_dir/ADNI/
data_dir/NACC/
data_dir/AIBL/
The tool was developped based on the following packages:
- PyTorch (1.1 or later)
- NumPy (1.16 or later)
- matplotlib (3.0.3 or later)
- tqdm (4.31 or later)
- matlab (2018b or later)
- scipy (1.4.1 or later)
Please note that the dependencies may require Python 3.6 or greater. It is recommemded to install and maintain all packages by using conda or pip. For the installation of GPU accelerated PyTorch, additional effort may be required. Please check the official websites of PyTorch and CUDA for detailed instructions.
The configuration file is a json file which allows you conveniently change hyperparameters of models used in this study.
{
"fcn":{
"fil_num": 20, # filter number of the first convolution layer in FCN
"drop_rate": 0.5,
"patch_size": 47, # 47 has to be fixed, otherwise the FCN model has to change accordingly
"batch_size": 10,
"balanced": 1, # to solve data imbalance issue, we provdided two solution: set value to 0 (weighted cross entropy loss), set value to 1 (pytorch sampler samples data with probability according to the category)
"Data_dir": "/data_dir/ADNI/", # change the path according to you folder name
"learning_rate": 0.0001,
"train_epochs": 3000
},
"mlp": {
"imbalan_ratio": 1.0, # imbalanced weight in weighted corss entropy loss
"fil_num": 100, # first dense layer's output size
"drop_rate": 0.5,
"batch_size": 8,
"balanced": 0,
"roi_threshold": 0.6,
"roi_count": 200,
"choice": "count", # if choice == 'count', then select top #roi_count as ROI
# if choice == 'thres', then select value > roi_threshold as ROI
"learning_rate": 0.01,
"train_epochs": 300
},
....
....
"cnn": {
"fil_num": 20,
"drop_rate": 0.137,
"batch_size": 2,
"balanced": 0,
"Data_dir": "/data/datasets/ADNI_NoBack/",
"learning_rate": 0.00001,
"train_epochs": 200
},
"cnnp": {
"fil_num": 20,
"drop_rate": 0.137,
"batch_size": 2,
"balanced": 0,
"Data_dir": "./ADNIP_NoBack/",
"learning_rate": 0.0001,
"train_epochs": 200
}
}{
"G_lr": 0.0066112,
"G_pth": "",
"G_fil_num": 10,
"warm_G_epoch": 5,
"D_lr": 0.01,
"D_pth": "",
"D_fil_num": 17,
"warm_D_epoch": 20,
"epochs": 5555,
"Data_dir": "/data/datasets/ADNI_NoBack/",
"L1_norm_factor": 0.1,
"AD_factor": 0.1,
"batch_size_p": 10,
"checkpoint_dir": "./checkpoint_dir/fcn_gan/",
"drop_rate": 0.725430,
"balanced": 0,
"log_name": "fcn_gan_log.txt",
"iqa_name": "iqa.txt",
"save_every_epoch": 30,
"D_G_ratio": 1
}python main.py
Please modify main.py to train and test your own models, where:
'gan_main' corresponding to training & generating phase of the GAN model, where one may choose a specified epoch for the generator & FCN.
'fcn_main' corresponding to training & testing phase of the FCN model.
'mlp_main' corresponding to training & testing phase of the multilayer perceptron model.
Function 'fcn_main' and 'mlp_main' will do number of repeat time independent FCN model training and MLP model on random splitted data. Model performance is thus evaluated on all runs as mean +/- std.
For the other functions, please explore them in the code & comments.
DPMs/fcn_exp0/
DPMs/fcn_exp1/
...
DPMs/fcn_expN/
Model weights and predicted raw scores on each subjects will be saved in:
ckeckpoint_dir/fcn_exp0/
ckeckpoint_dir/fcn_exp1/
...
ckeckpoint_dir/fcn_expN/