Mark Akeson (1), Andrew D. Beggs (2), Thomas Nieto (2), Miten Jain (1), Nicholas J. Loman (3), Matt Loose (4), Sunir Malla (4), Justin O’Grady (5), Hugh E. Olsen (1), Josh Quick (3), Hollian Richardson (5), Jared T. Simpson (6,7), Terrance P. Snutch (8), Louise Tee (2), John R. Tyson (8)
- University of California, Santa Cruz, Santa Cruz, CA, USA
- University of Birmingham, Birmingham, B15 2TT
- Institute of Microbiology and Infection, School of Biosciences, University of Birmingham, Birmingham, B15 2TT, United Kingdom
- DeepSeq, School of Life Sciences, University of Nottingham, Nottingham, UK
- Norwich Medical School, University of East Anglia, Norwich, NR4 7UQ, United Kingdom.
- Ontario Institute for Cancer Research, Toronto, Canada
- Department of Computer Science, University of Toronto, Toronto, Canada
- Michael Smith Laboratories, University of British Columbia, Vancouver, Canada
We have sequenced the CEPH1463 (NA12878/GM12878, Ceph/Utah pedigree) human genome reference standard on the Oxford Nanopore MinION using 1D ligation kits (450 bp/s) using R9.4 chemistry (FLO-MIN106).
Human genomic DNA from GM12878 human cell line (Ceph/Utah pedigree) was either purchased from Coriell - "DNA" - (cat no NA12878) or extracted from the cultured cell line - "cells". As the DNA is native, modified bases will be preserved.
Check back in the next few days for the remaining reads, alignments and raw signal-level reads.
We have processed approximately 2/3rds of the total dataset.
The current release rel2 consists of:
- 25 flowcells
- 58958035887 bases
- 9053909 reads
| flowcell_id | reads | bases | Date | Centre | SampleType | Links | Link |
|---|---|---|---|---|---|---|---|
| FAB39075 | 466324 | 2439308482 | 20/09/16 | UBC | DNA | FASTQ | BAM |
| FAB39043 | 305667 | 1543725551 | 23/09/16 | Bham | DNA | FASTQ | BAM |
| FAB42706 | 400751 | 1857323339 | 12/10/16 | UBC | DNA | FASTQ | BAM |
| FAB42316 | 107013 | 606761274 | 14/10/16 | Notts | DNA | FASTQ | BAM |
| FAB42205 | 312666 | 1664297400 | 14/10/16 | Notts | DNA | FASTQ | BAM |
| FAB42561 | 231562 | 1510037000 | 19/10/16 | Notts | DNA | FASTQ | BAM |
| FAB42473 | 598480 | 3140575707 | 20/10/16 | UBC | DNA | FASTQ | BAM |
| FAB42476 | 376897 | 2061807133 | 27/10/16 | UBC | DNA | FASTQ | BAM |
| FAB42451 | 769524 | 4256154457 | 28/10/16 | Notts | DNA | FASTQ | BAM |
| FAB42704 | 276151 | 1750146174 | 28/10/16 | UBC | DNA | FASTQ | BAM |
| FAB42810 | 265456 | 1665251718 | 02/11/16 | Norwich | DNA | FASTQ | BAM |
| FAB46683 | 72602 | 286264094 | 17/11/16 | Bham | DNA | FASTQ | BAM |
| FAB45332 | 530913 | 2863965040 | 17/11/16 | UBC | DNA | FASTQ | BAM |
| FAB43577 | 241646 | 1423672212 | 18/11/16 | UCSC | DNA | FASTQ | BAM |
| FAB44989 | 558195 | 3443623448 | 18/11/16 | UCSC | DNA | FASTQ | BAM |
| FAF01169 | 16489 | 120873419 | 22/11/16 | Bham | Cells | FASTQ | BAM |
| FAF01441 | 43281 | 358912895 | 22/11/16 | Bham | Cells | FASTQ | BAM |
| FAB45277 | 53541 | 445614920 | 22/11/16 | Notts | Cells | FASTQ | BAM |
| FAB45321 | 299172 | 2583989736 | 22/11/16 | Notts | Cells | FASTQ | BAM |
| FAF01132 | 689781 | 5455971336 | 25/11/16 | Bham | Cells | FASTQ | BAM |
| FAF01127 | 632728 | 4972081712 | 25/11/16 | Bham | Cells | FASTQ | BAM |
| FAB49712 | 592317 | 4589575564 | 28/11/16 | Bham | Cells | FASTQ | BAM |
| FAF01253 | 442221 | 3476220233 | 28/11/16 | Bham | Cells | FASTQ | BAM |
| FAB49914 | 309162 | 2840857895 | 28/11/16 | Notts | Cells | FASTQ | BAM |
| FAB45271 | 461370 | 3601025148 | 28/11/16 | Notts | Cells | FASTQ | BAM |
Please verify downloads against MD5 hashes and list of links.
Reads aligned against pre-computed 1000 genomes GRCh38 BWA database at ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/reference/GRCh38_reference_genome/ with decoys using BWA MEM (commit: 5961611c358e480110793bbf241523a3cfac049b) using parameters -x ont2d. Alignment statistics calculated using samtools stats (samtools version 1.3.1).
| FlowcellID | Sequences | Mapped | Mapped.MQ0 | Unmapped | Bases.Mapped | Avg.Length | Link |
|---|---|---|---|---|---|---|---|
| FAB39075 | 466325 | 425113 | 28167 | 41212 | 2146410660 | 5230 | BAM |
| FAB39043 | 305667 | 294755 | 13692 | 10912 | 1457990655 | 5050 | BAM |
| FAB42706 | 400751 | 353625 | 15675 | 47126 | 1776803301 | 4634 | BAM |
| FAB42316 | 107013 | 95753 | 3137 | 11260 | 586781780 | 5669 | BAM |
| FAB42205 | 312666 | 278495 | 12223 | 34171 | 1582844105 | 5322 | BAM |
| FAB42561 | 231562 | 223447 | 10030 | 8115 | 1412971869 | 6521 | BAM |
| FAB42473 | 598480 | 571453 | 28466 | 27027 | 2929628858 | 5247 | BAM |
| FAB42476 | 376897 | 364557 | 17046 | 12340 | 1953918702 | 5470 | BAM |
| FAB42451 | 769524 | 738272 | 33281 | 31252 | 3963057878 | 5530 | BAM |
| FAB42704 | 276151 | 263721 | 12926 | 12430 | 1619871937 | 6337 | BAM |
| FAB42810 | 265456 | 251084 | 14164 | 14372 | 1483597856 | 6273 | BAM |
| FAB46683 | 72602 | 64736 | 5306 | 7866 | 269203835 | 3942 | BAM |
| FAB45332 | 530913 | 497838 | 26391 | 33075 | 2620591926 | 5394 | BAM |
| FAB43577 | 241646 | 231695 | 11028 | 9951 | 1321809290 | 5891 | BAM |
| FAB44989 | 558195 | 536543 | 25934 | 21652 | 3161714885 | 6169 | BAM |
| FAF01169 | 16489 | 15772 | 710 | 717 | 114528983 | 7330 | BAM |
| FAF01441 | 43281 | 41775 | 1847 | 1506 | 335717952 | 8292 | BAM |
| FAB45277 | 53541 | 51951 | 2132 | 1590 | 426614901 | 8322 | BAM |
| FAB45321 | 299172 | 283353 | 15165 | 15819 | 2365977206 | 8637 | BAM |
| FAF01132 | 689781 | 655357 | 33564 | 34424 | 4966810089 | 7909 | BAM |
| FAF01127 | 632728 | 605633 | 27192 | 27095 | 4640355789 | 7858 | BAM |
| FAB49712 | 592317 | 576236 | 24321 | 16081 | 4308054580 | 7748 | BAM |
| FAF01253 | 442221 | 428640 | 18700 | 13581 | 3245588740 | 7860 | BAM |
| FAB49914 | 309162 | 296238 | 12280 | 12924 | 2673699794 | 9188 | BAM |
| FAB45271 | 461370 | 440262 | 19535 | 21108 | 3388203702 | 7805 | BAM |
Please verify downloads against MD5 hashes.
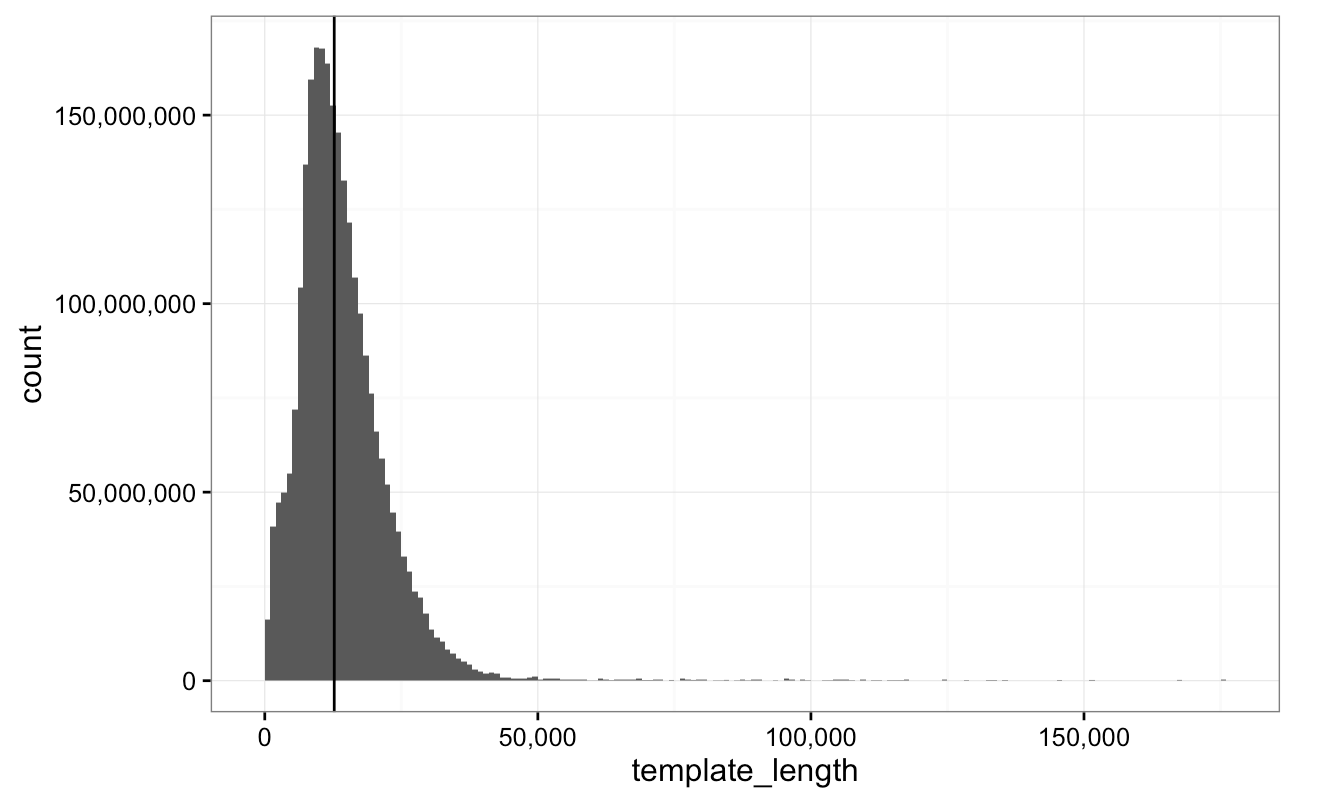
Figure: A typical read length distribution from a flowcell where we have run a cell-extracted DNA library. The y-axis shows the count of bases. Mean read length ~8.6kb with N50 of ~12.5kb (vertical line). Reads longer than 60kb are not expected due to limitations of the QIAGEN extraction kit employed.
This dataset is currently subject to rapid change as we continue to post up runs, therefore some statistics here may not represent full nanopore runs.
We would like to acknowledge the support of Oxford Nanopore Technologies in generating this dataset, with particular thanks to Rosemary Dokos, Oliver Hartwell, Jonathan Pugh and Clive Brown. We would like to thank Radoslaw Poplawski and Simon Thompson for technical assistance with configuration and optimising of the CLIMB platform file system.
Please raise issues on this Github repository concerning this dataset. A preprint describing the dataset in more detail will be available shortly.
* rel1: 1st December 2016. Initial release.