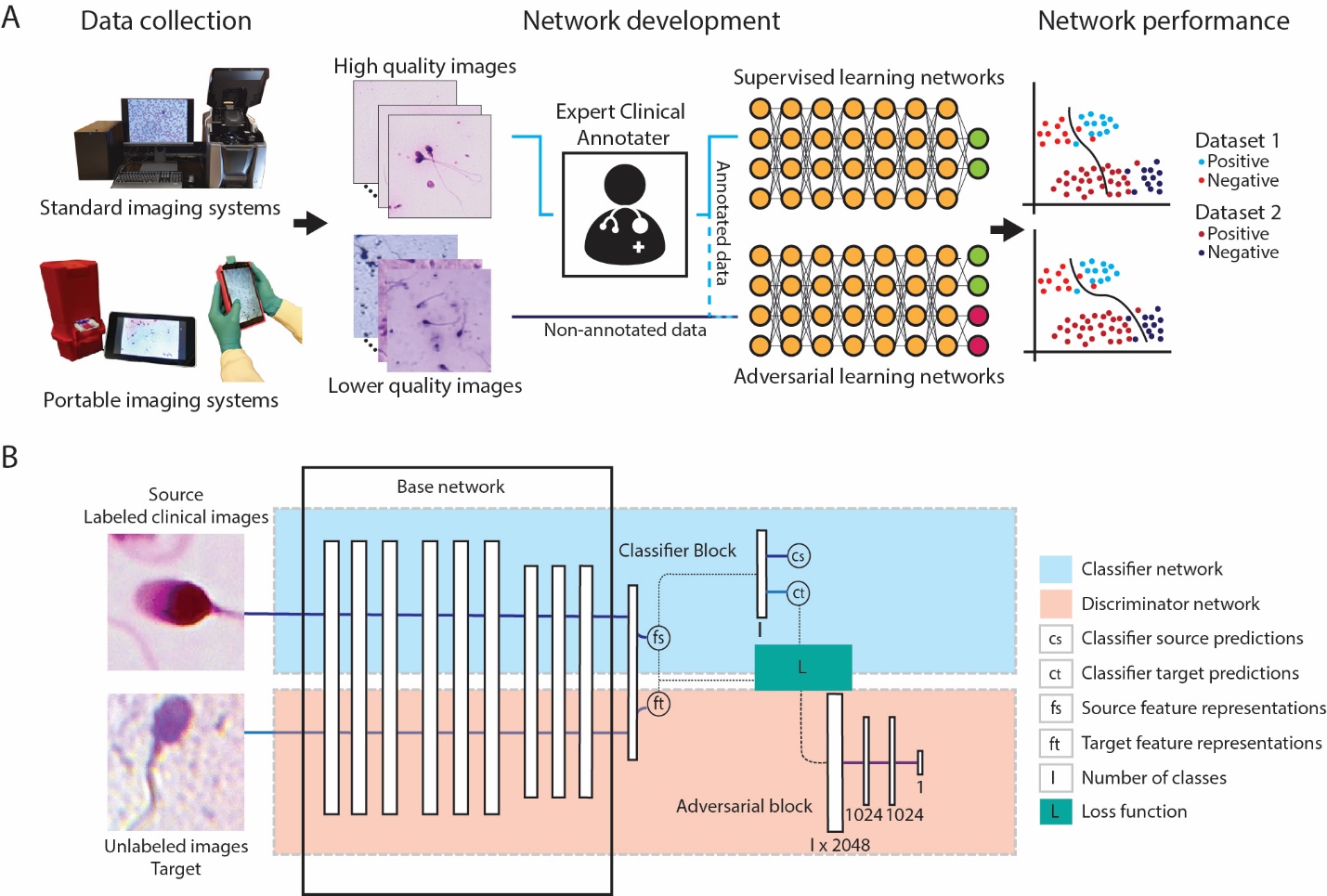
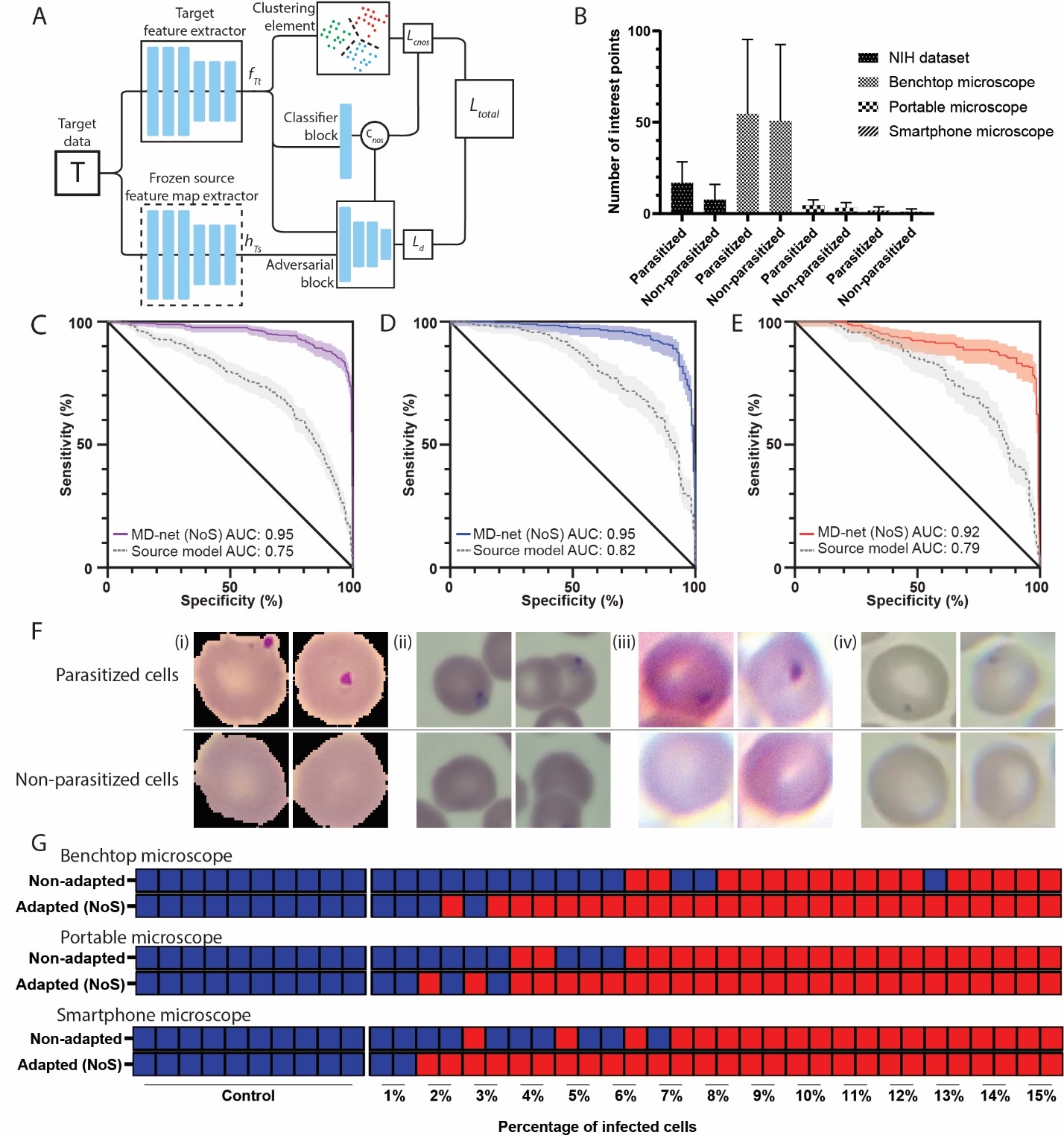
This is the Pytorch implementation for our paper Adaptive adversarial neural networks for the analysis of lossy and domain-shifted datasets of medical images, Nature Biomedical Engineering
TLDR; Adversarial learning can be used to develop high-performing networks trained on unannotated medical images of varying image quality, and to adapt pretrained supervised networks to new domain-shifted datasets.
In machine learning for image-based medical diagnostics, supervised convolutional neural networks are typically trained with large and expertly annotated datasets obtained using high-resolution imaging systems. Moreover, the network’s performance can degrade substantially when applied to a dataset with a different distribution. Here, we show that adversarial learning can be used to develop high-performing networks trained on unannotated medical images of varying image quality. Specifically, we used low-quality images acquired using inexpensive portable optical systems to train networks for the evaluation of human embryos, the quantification of human sperm morphology and the diagnosis of malarial infections in the blood, and show that the networks performed well across different data distributions. We also show that adversarial learning can be used with unlabelled data from unseen domain-shifted datasets to adapt pretrained supervised networks to new distributions, even when data from the original distribution are not available. Adaptive adversarial networks may expand the use of validated neural-network models for the evaluation of data collected from multiple imaging systems of varying quality without compromising the knowledge stored in the network.
- Linux (Tested on Ubuntu 18.04.05)
- NVIDIA GPU (Tested on Nvidia GeForce GTX 1080 Ti x 4 on local workstations, and Nvidia V100 GPUs on Cloud)
- Python (Tested with v3.6)
- Pytorch 1.4.0
- PyYAML 5.3.1
- scikit-image 0.14.0
- scikit-learn 0.20.0
- SciPy 1.1.0
- opencv-python 4.2.0.34
- Matplotlib 3.0.0
- NumPy 1.15.2
.txt files are lists for source and target domains
The Embryo, Malaria, Sperm are available online here and Office-31 datasets from here Once they are downloaded and extracted into your data directory, create .TXT files with filepaths and numeric annotation with space delimited.
The data used for training and testing are suggested to be organized as follows:
DATA_ROOT_DIR/
└── DATA_SET_NAME
├── DISTRIBUTION_SET_NAME
├── CLASS_NUMBER
. ├── file_name.png
. └── file_name.png
. .
├── CLASS_NUMBER
. ├── file_name.png
. └── file_name.png
└── DISTRIBUTION_SET_NAME
├── CLASS_NUMBER
. ├── file_name.png
. └── file_name.png
. .
├── CLASS_NUMBER
. ├── file_name.png
. └── file_name.png| D | [ S-Sperm, E-Embryo, M-Malaria/Redblood cells ] | |
| DYst | Y | [ M-Model, D-Dataset] |
| s | [data distribution] | |
| t | [data distribution] |
For dataset and imaging system nomenclature, s is graded from 4 through 1, with 4 being the dataset/imaging system of highest quality image (clinical microscope systems) and 1 being the dataset/imaging system of lowest quality (smartphone microscope systems). For example, ED4 denotes the embryo dataset imaged using a clinical Embryoscope that was used by embryologists for the annotations.
t is only used when defining domain adaption models, to denote source (s) and target (t) that were used when developing the model. For example, EM41 denotes a model (M) trained on embryo datasets (E) with Embryoscope images (4) as its source and smartphone images (1) as its target.
You can train MD-nets as follows
python -u experiments/python_scripts/train_md_nets.py --mode train \
--seed seed
--num_iterations 100000 --patience 5000 --test_interval 500 --snapshot_interval 1000 \
--dset dataset_name --s_dset source_name --t_dset target_name \
--lr "0.0001" --batch_size 32 --optimizer SGD --use_bottleneck true --batch_size_test 256
--s_dset_txt "source_training.txt"
--sv_dset_txt "source_validation.txt" \
--t_dset_txt "target.txt" \
--no_of_classes 5 \
--output_dir "experiments" \
--gpu_id 1 \
--arch Xception\
--crop_size 224 --image_size 256
--target_labelled true --trade_off 1.0 \
--trained_model_path ""python -u experiments/python_scripts/train_md_nets_nos.py --mode train \
--seed seed
--num_iterations 10000 --patience 5000 --test_interval 500 --snapshot_interval 1000 \
--dset embryo --s_dset source_name --t_dset target_name \
--lr "0.0001" --batch_size 32 --optimizer SGD --use_bottleneck true --batch_size_test 256
--tv_dset_txt "target_validation.txt" \
--t_dset_txt "target.txt" \
--no_of_classes 31 \
--output_dir "experiments" \
--gpu_id 0 \
--arch ResNet50\
--crop_size 224 --image_size 256
--source_model_path "model.pth.tar"
--trade_off_cls 0.3 To run all the experiments reported in the paper
./experiments/scripts/run_DATA_SET_NAME_experiments.sh
The experiment log file and the saved models will be stored at ./experiments/logs/EXPERIMENT_NAME/ and ./experiments/models/EXPERIMENT_NAME
You can test the datasets on reported models as follows
./experiments/scripts/test_DATASETNAME.sh
If you find our work useful in your research or if you use parts of this code please consider citing our paper in your research:
@article{Kanakasabapathy2021,
doi = {10.1038/s41551-021-00733-w},
url = {https://doi.org/10.1038/s41551-021-00733-w},
year = {2021},
month = jun,
publisher = {Springer Science and Business Media {LLC}},
author = {Manoj Kumar Kanakasabapathy and Prudhvi Thirumalaraju and Hemanth Kandula and Fenil Doshi and Anjali Devi Sivakumar and Deeksha Kartik and Raghav Gupta and Rohan Pooniwala and John A. Branda and Athe M. Tsibris and Daniel R. Kuritzkes and John C. Petrozza and Charles L. Bormann and Hadi Shafiee},
title = {Adaptive adversarial neural networks for the analysis of lossy and domain-shifted datasets of medical images},
journal = {Nature Biomedical Engineering}
}
© Shafiee Lab - This code is made available under the GNU GPLv3 License and is available for non-commercial academic purposes.
If you have any questions, please contact us via hshafiee[at]bwh.harvard.edu.