mysoftware
This is the software repository for our paper solving problem x in a novel way.
Repository layout (what do I find where?)
|- container # OCI recipes
|- mymodule # source code for this software module
|- images
|- notes # Project organization, paper LaTex, related work, proposal, meeting notes
|- scripts # processing scripts, standalone CLI
Motivation
Short description of
- problem in field (e.g. cancer)
- what is current solution (handdrawn segmentation)
- what this software does to solve it (sota segmentation)
- proposed impact (accelerated/accurate diagnosis) Similar to abstract of the paper
Status
Table of contents
Installation
Local
git clone <thisrepo>
# build instructionsPip
pip install mysoftwareConda
conda install mysoftwareor using the environment
conda env create --name myproject --file=myproject.ymlContainer
Singularity
Building from recipes
sudo singularity build container/singularityrecipe.defReleased images
singularity pull repo:image.versionTest
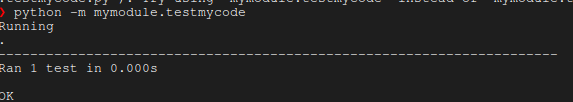
To run the tests ensuring the installation is valid, please run
python -m mymodule.testmycode
Which should result in
Usage
Example snippets
Example inference on in silico data
import seaborn as sns
import mysoftware.preprocessing as stk
import numpy as np
np.random.seed(42)
silico_data = stk.generate_insilico(args)
q = stk.infer(silico_data)
sns.boxplot(x='method', y='accuracy', data=q)Notebooks
See notebooks/example.ipynb for an example interactive workflow.
API
See docs for the complete API documentation
Cite
@article{you2023,
title={yourtitle},
author={authors et al},
year={2023},
publisher={venue}
}
Reproducing our published results
Data
In silico
See [mysoftware/insilico] on how to generate our in silico datasets. For your convenience, this can be downloaded as well:
mkdir insilico && cd insilico
wget -O insilico.hdf5 https://mydatarepo/insilico.hdf5Real world
mkdir mydata
wget -O dataset.hdf5 https://mydatarepo/dataset.hdf5Ensure the checksums match
md5sum dataset.hdf5should produce
4a4f224c7b7c871855fd307ae323be93 dataset.hdf5Pre/post processing code
See scripts/preprocessing.py for the preprocessing scripts that configure the dataset and generate the plots
FAQ
Help I can't figure out how function5 works
Please create a new issue detailing concisely, yet complete what issue you encountered, in a reproducible way.
FOR STUDENTS -- REMOVE ME
This a template for you to reuse in releasing software accompanying papers. At a minimum, ensure your software runs in a reproducible way. Place your (Python) code in [mymodule/mycode.py] (or different submodules). Processing scripts ideally are separate from the modules
Instructions on how to create packages/containers: