ERGO: Efficient Recurrent Graph Optimized Emitter Density Estimation in Single Molecule Localization Microscopy
This repository holds the source accompanying our IEEE Transactions in Medical Imaging 2020 paper.
Repository organization
ERGO has 2 stages:
This repository holds the Python code for the emitter density prediciton stage (ER).
All files in this repository are licensed under Affero GPL v 3, copyright 2018-2021 Ben Cardoen. The software was developed in a multidisciplinary collaboration between the labs of Prof. Ghassan Hamarneh, Prof. Ivan Robert Nabi, and Prof. Keng C. Chou.
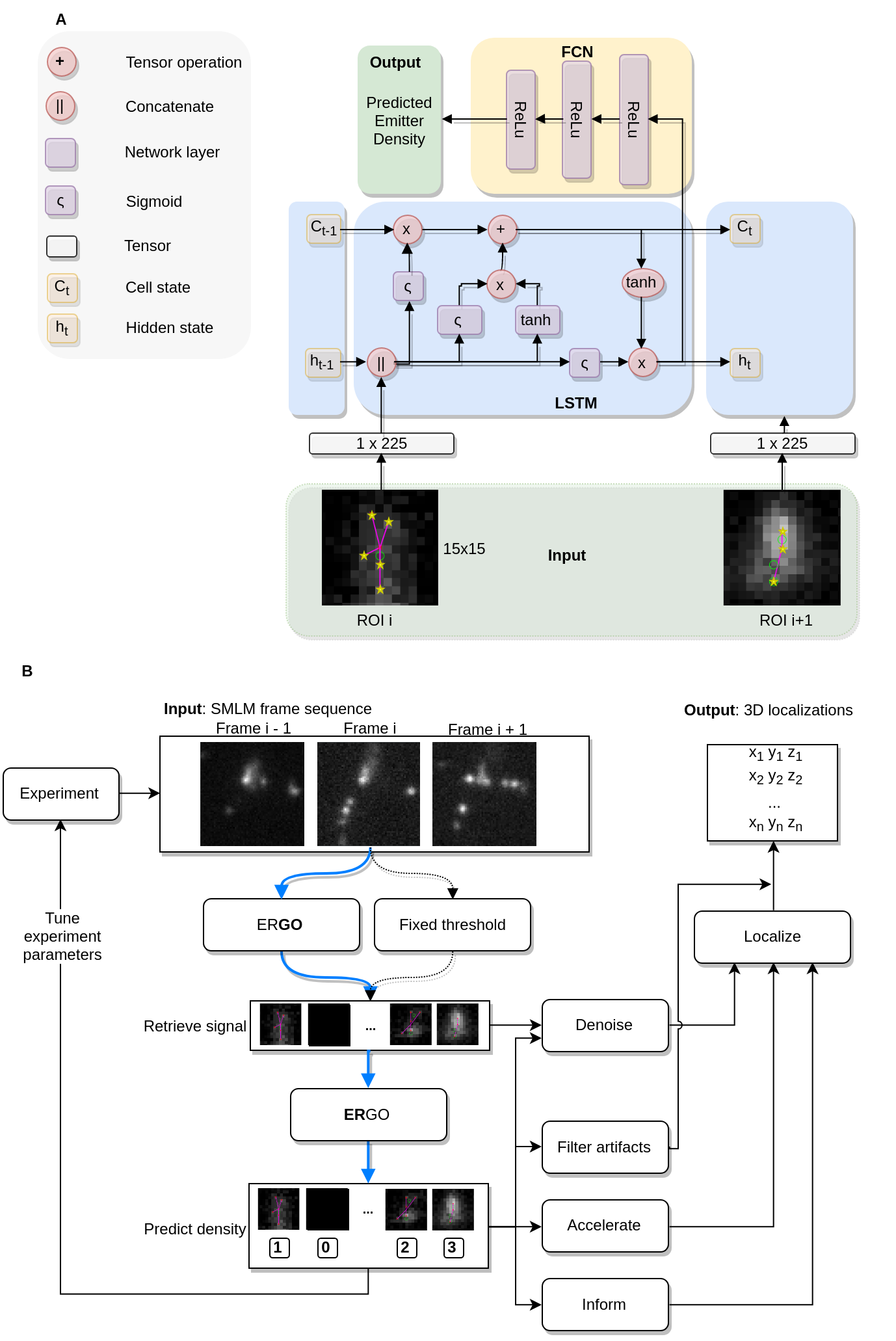
We refer to the manuscript for a detailed explanation of architecture choices, losses, configuration, and so on. Summarized, we leverage the information present in the stream of ROIs from at most 4 consecutive frames to more accurately predict density. Given that the frame sequence is highly variable, but only strongly correlates across a few consecutive frames, we enable specialization without overfitting. The emission density per ROI (and frame) follows a Poisson distribution, learning such a counting process is non-trivial. To ensure we capture also the rare (high density) events we weight our loss accordingly. The manuscript details this in detail.
Layout
- output
- net : saved network(s)
- example results on training data are also included as figures
- see src/plotter.py for plotted output
- src : python code + SLURM submissions scripts
- trainlstm.py
- data : optional training data (images + csv with density GT), you can generate this using the ERGO.jl package, or use the data provided for your convenience.
- training.tgz, which when extracted should offer:
- frame_x_y.tif
- ROI nr y from frame x
- density.csv
- framenr roinr mx my count : mx/my are anchor locations in frame of ROI, count is GT of nr emissions in this ROI
- frame_x_y.tif
- see https://github.com/bencardoen/ERGO.jl on how to create similar training data from different sources
- training.tgz, which when extracted should offer:
Installation - Dependencies
- venv: see requirements_cedar.txt (compatible with Compute Canada) or requirements_local.txt
- conda : see environment.yml (change the location directory)
For example, using conda you would do:
conda env create --name ERGO --file=environment.yml
conda activate ERGOUsage
This project was trained on SLURM (HPC) based systems (e.g. Compute Canada), but ran equally well on my laptop (with 4G GPU). An example submission script is added in ./src, but you can run the code also in a standard command line.
Please see src/submit.sh for an example on how to run the training/evaluation code. The arguments are documented in trainlstm.py
First, we extract data. Assume we start from repository directory.
cd data
tar -xf training.tgzThen
conda activate ERGO
cd src
python trainlstm.py --imageroot=../data --loss=intmse --outputroot=/dev/shm/output --shuffle=False --mode=lstm --epochs=500 --csv=../data/density.csv --mode=validate --batchweights=TrueThis would look something like the below
The trainlstm.py script can do both training or evaluation only (using a saved network).
A summary listing of the parameters: (defaults are set in trainlstm.py line 33)
- imageroot: directory with ROIs in tiff format
- outputroot : where to save produced data
- shuffle : randomize input (not a great idea for LSTM)
- mode : LSTM, of FCN (e.g. ignore temporal data)
- epochs : self explanatory
- csv : where to find GT CSV data
- batch : batchsize (64)
- loss : which loss function to use
- pix : pixel size of 1`ROI
- logdir : where to write the full log file
- mode : train/test/validate/evaluate
- initstrategy : zero/random, for the weights
- seed : default 1
- lr : learning rate. Note that we use an oscillating LR
- trimclasses : if ROI x has k emitters, and trimclasses is k-2, bin all densities > k-2 into k-2. Useful for extreme long tail data. Not needed usually
- batchweights : off, compute weights (frequency) per batch to adapt to highly variable Poisson like data (recommended, but slow)
- pretraineddir : where to find saved network, only used if mode = evaluate. For examples, see ./output/net/save_me/*.pth
FAQ
- I want to use this for my own data, how do I do this ?
- Check https://github.com/bencardoen/ERGO.jl/blob/main/README.md on how to generate data in the format ER expects, an example notebook is provided.
If you use this code, please cite
@article{cardoen2019ergo,
title={Ergo: efficient recurrent graph optimized emitter density estimation in single molecule localization microscopy},
author={Cardoen, Ben and Yedder, Hanene Ben and Sharma, Anmol and Chou, Keng C and Nabi, Ivan Robert and Hamarneh, Ghassan},
journal={IEEE transactions on medical imaging},
volume={39},
number={6},
pages={1942--1956},
year={2019},
publisher={IEEE}
}